Exporting Sequences to Sequence Format
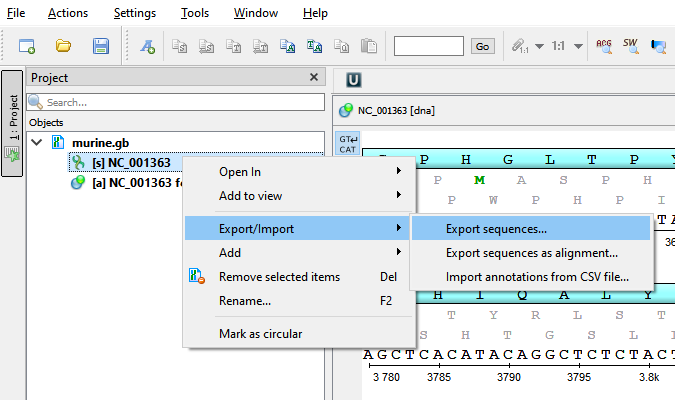
Select one or several sequence objects in the Project View window and click the Export/Import ‣ Export sequences context menu item:
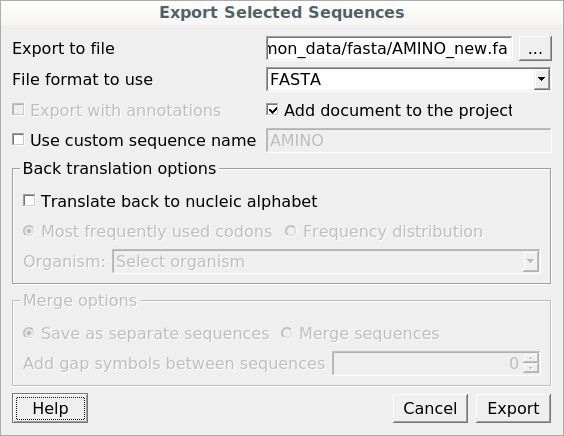
The Export Selected Sequences dialog will appear for nucleotide sequences:
The Export Selected Sequences dialog will appear for amino acid sequences:
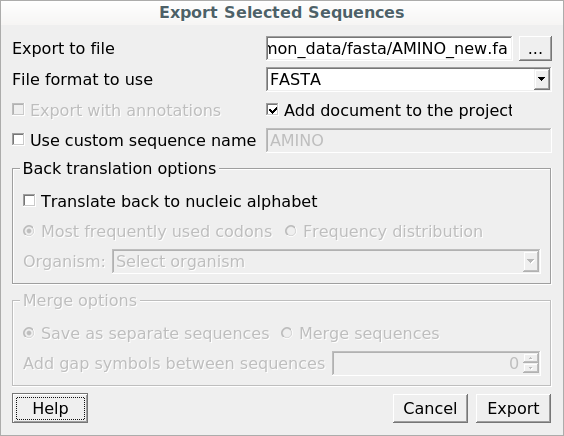
In this dialog, you can select the location of the output file and choose a sequence file format. You also have the option to add the newly created document to the current project and use a custom sequence name. To do so, check the corresponding checkboxes.
Use the Conversion options to choose a strand for saving the sequence(s). You can also translate the sequence(s) to an amino/nucleic alphabet.
Additionally, it is possible to specify whether to merge the exported sequences into a single sequence or store them as separate sequences. If you merge the sequences, you’re allowed to select the gap symbols between sequences. These symbols define the length of the insertion region between sequences, containing N symbols for nucleic or X for protein sequences.
Export sequence with annotations
To export a sequence with annotations, choose the Genbank or GFF format. The Export with annotations checkbox will be available. Check the checkbox, and the sequence will be exported with annotations.