Fetching Data from Remote Database
UGENE allows the fetching of data from remote biological databases such as NCBI GenBank, the NCBI protein sequence database, and others.
To fetch data, select the File ‣ Access remote database… item in the main menu.
The dialog will appear:
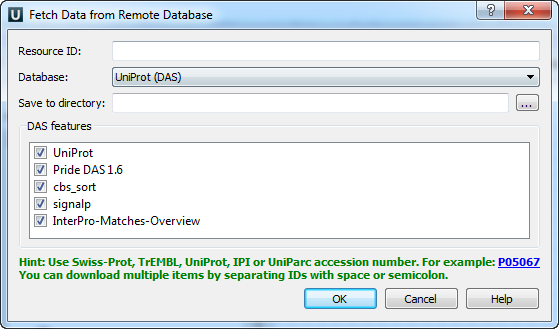
The following parameters are available:
Resource ID: Enter the unique ID of the biological object and choose a database. Unique identifiers differ for various databases. For example, for NCBI GenBank such a unique ID could be an Accession Number or an NCBI GI number.
Database: The following databases are available: NCBI GenBank (DNA sequence), NCBI protein sequence database, ENSEMBL, PDB, SWISS-PROT, UniProtKB/Swiss-Prot, UniProtKB/TrEMBL. You can open the database page by clicking the arrow button at the end
 where the link to the external database is stored.
where the link to the external database is stored.Save to directory: Browse for a directory to save the fetched file.
Add to project: Select this checkbox if you want to add the document to a project.
Force download the appropriate sequence: Some entries in NCBI databases contain features without sequences. You can download both sequences and features by clicking this parameter. Note that some sequences are rather large. For example, only annotations without sequences will be loaded for NC.000853.1 if this parameter is unchecked.
After you click the OK button, UGENE downloads the biological object (DNA sequence, protein sequence, 3D model, etc.) and adds it to the current project.
A task report with the specified information will be generated after downloading.
If something goes wrong, check the Log View; it will help you diagnose the problem.