Searching NCBI Genbank
UGENE enables users to search data in the NCBI GenBank remote database. To do this, open the following dialog through the main menu by selecting File->Search NCBI Genbank:
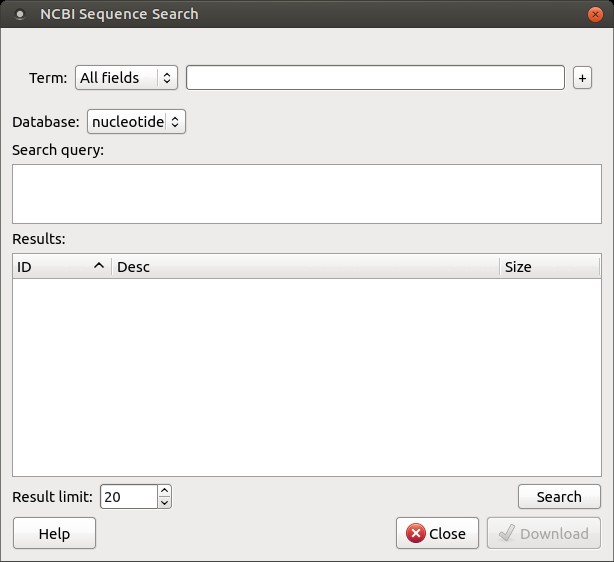
To search for data in the nucleotide or protein databases, enter a general text query in the search field, select the appropriate database, and click the Search button. You can use a protein name, gene name, or gene symbol directly. Searching for a submitter or author name in the specific format will yield the best results.
Use the boolean operator AND to find records containing all your search terms, which is the intersection of search results.
Use the boolean operator OR to find records that include any of several search terms, which is the union of search results.
Use the boolean operator NOT to exclude records matching a particular search term.
To limit the results, use the Result limit field.
After clicking the Search button, UGENE searches for the biological objects and displays them in the Results field. You can download the object(s) by selecting one or more objects (to select multiple objects, use the Ctrl button) and clicking the Download button. The dialog will appear:
The meanings of all parameters are similar to those in the dialog Fetch Data from Remote Database, which is accessed through File→Access Remote Database.
You can save the downloaded file in one of two formats: fasta or GenBank.
The Force download appropriate sequence parameter is not available if you select the fasta format.
After clicking the OK button, UGENE downloads the biological objects and adds them to the current project if the Add to project option is checked.