Making Request to Database
To make a request to a local BLAST database, do the following:
- Open Tools ‣ BLAST ‣ BLAST search.
If there is a sequence opened, you can also initiate the request to a local BLAST database from the Sequence View:
- Select the Analyze ‣ Query with local BLAST item in the context menu or in the Actions menu in the main menu.
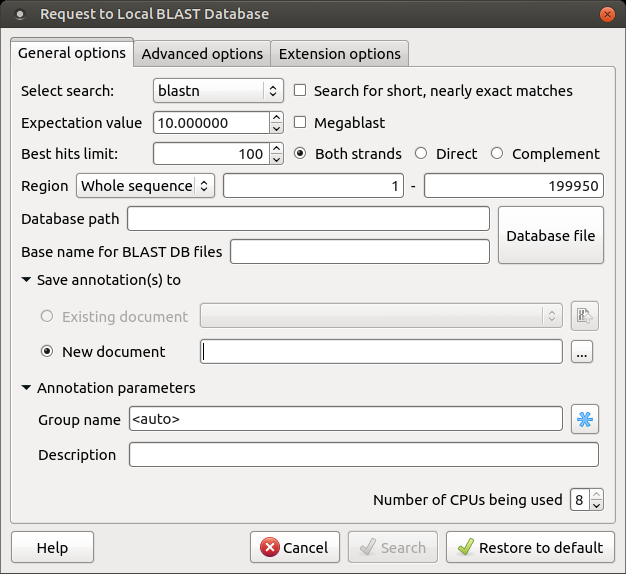
The Request to local BLAST database dialog will appear:
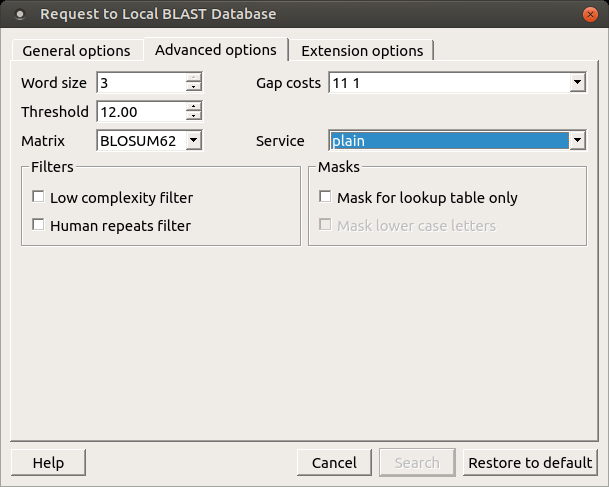
The following general options are available:
- Select search: Here you should select the tool you would like to use. If the query sequence is a nucleotide sequence, then blastn, blastx, and tblastx items are available. For a protein sequence, the items are blastp and tblastn.
- Expectation value: This option specifies the statistical significance threshold for reporting matches against database sequences. Lower expect thresholds are more stringent, leading to fewer chance matches being reported.
- Culling limit: The maximum number of hits that will be shown (not equal to the number of annotations). The maximum available number is 5000.
- Search for short, nearly exact matches: Automatically adjusts the word size and other parameters to improve results for short queries.
- Megablast: Select this option to compare queries with closely related sequences. It works best if the target percent identity is 95% or more, but it is very fast.
- Database path: Path to the database files.
- Base name for BLAST DB files: The base name for the BLAST database files.
You can see the description of the annotation saving parameters here.
The following advanced parameters are available:
- Word size: The size of the subsequence parameter for the initiated search.
- Gap costs: Costs to create and extend a gap in an alignment. Increasing the gap costs will result in alignments that decrease the number of gaps introduced.
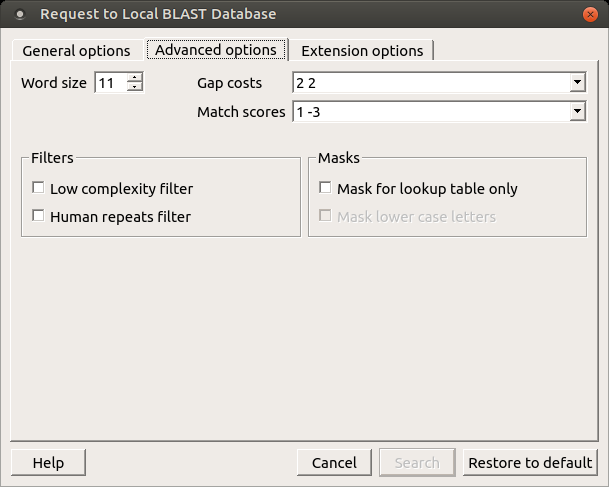
- Match scores: Reward and penalty for matching and mismatching bases.
- Filters: Filters for regions of low compositional complexity and repeat elements of the human genome.
- Masks for lookup table only: This option masks only for purposes of constructing the lookup table used by BLAST so that no hits are found based upon low-complexity sequences or repeats (if repeat filter is checked).
- Mask lower case letters: With this option selected, you can cut and paste a FASTA sequence in uppercase characters and denote areas you would like filtered with lowercase.
The view of the Advanced options tab depends on the selected search. For a blastn search, it looks like the picture above. When the blastx search is selected in the general options, the view of the Advanced options tab is as follows:
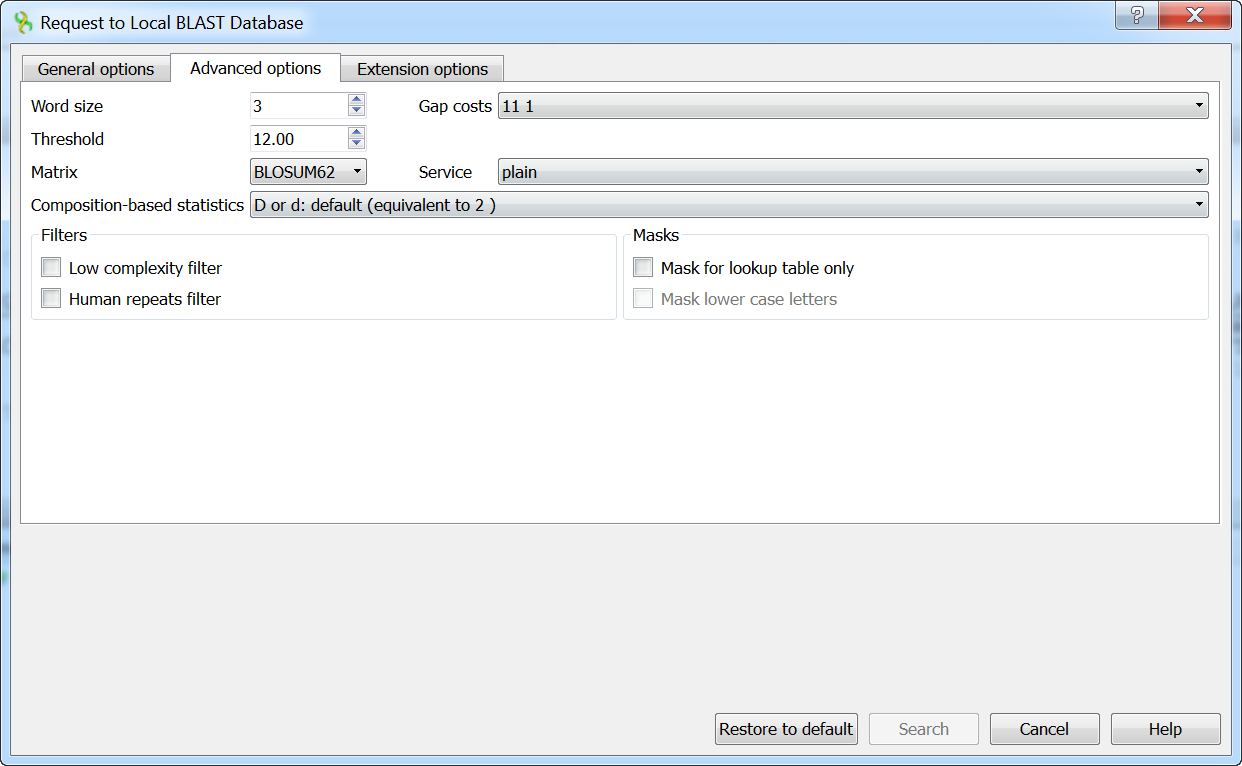
As you can see, there is no Match scores option, but there are Threshold, Matrix, Composition-based statistics, and Service options.
- Threshold: Threshold for extending hits.
- Matrix: The key element in evaluating the quality of pair-wise sequence alignment is the “substitution matrix,” which assigns a score for aligning any possible pair of residues.
- Service: The blastp service that needs to be performed: plain, psi, or phi.
- Composition-based statistics: Composition-based statistics.
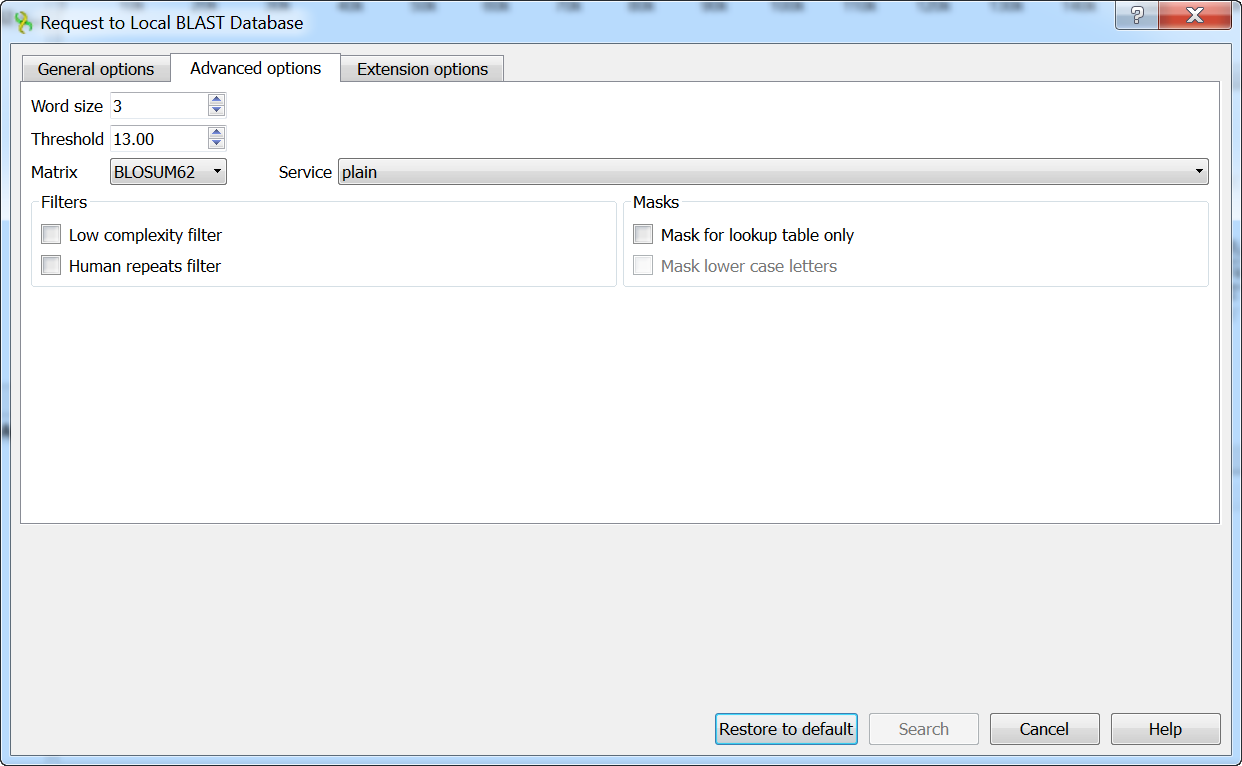
When the tblastx search is selected in the general options, the view of the Advanced options tab is as follows:
The following extension options are available:
- For gapped alignment: X dropoff value (in bits) for gapped alignment.
- For ungapped alignment: X dropoff value (in bits) for ungapped alignment.
- For final gapped alignment: X dropoff value (in bits) for final gapped alignment.
- Multiple hits window size: Multiple hits window size.
- Perform gapped alignment: Performs gapped alignment.