Aligning Short Reads with BWA-SW
When you select the Tools ‣ Align to reference ‣ Align short reads option in the main menu, the Align Sequencing Reads dialog box will appear. Set the value of the Align short reads method parameter to BWA-SW. The dialog box looks as follows:
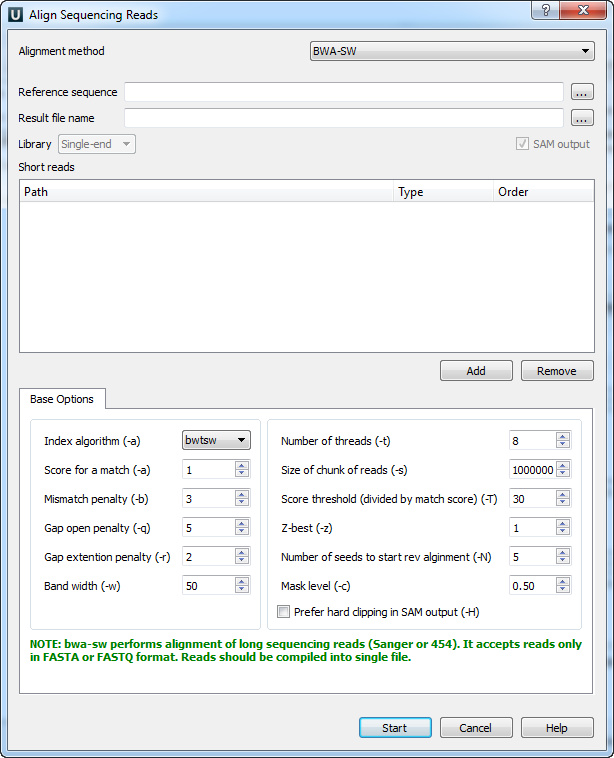
The following parameters are available:
Reference sequence — The DNA sequence to align short reads to. This parameter is required.
Result file name — The file in SAM format to write the result of the alignment into. This parameter is required.
SAM output — Always save the output file in the SAM format (this option is disabled for BWA).
Short reads — Each added short read is a small DNA sequence file. At least one read should be added.
You can also configure other parameters:
Index algorithm (-a) — The algorithm for constructing the BWA-SW index. It implements three different algorithms:
- is — Designed for short reads up to ~200bp with a low error rate (<3%). It performs gapped global alignment with respect to reads, supports paired-end reads, and is one of the fastest short read alignment algorithms to date while also visiting suboptimal hits.
- bwtsw — Designed for long reads with more errors. It performs heuristic Smith-Waterman-like alignment to find high-scoring local hits. The algorithm is implemented in BWA-SW. On low-error short queries, BWA-SW is slower and less accurate than the is algorithm, but on long reads, it is better.
- div — Not suitable for long genomes.
Score for a match (-a) — The score of a match.
Mismatch penalty (-b) — The mismatch penalty.
Gap open penalty (-q) — The gap open penalty.
Gap extension penalty (-r) — The gap extension penalty. The penalty for a contiguous gap of size k is q+k*r.
Band width (-w) — The band width in the banded alignment.
Number of threads (-t) — The number of threads in multi-threading mode.
Size of chunk of reads (-s) — The maximum SA interval size for initiating a seed. A higher -s increases accuracy at the cost of speed.
Score threshold (divided by match score) (-T) — The minimum score threshold.
Z-best (-z) — Z-best heuristics. A higher -z increases accuracy at the cost of speed.
Number of seeds to start reverse alignment (-N) — The minimum number of seeds supporting the resultant alignment to skip reverse alignment.
Mask level (-c) — Coefficient for threshold adjustment according to query length. Given an l-long query, the threshold for a hit to be retained is a*max{T,c*log(l)}.
Prefer hard clipping in SAM output (-H) — Use hard clipping in the SAM output. This option may dramatically reduce redundancy in the output when mapping long contig or BAC sequences.
Select the required parameters and press the Start button.