ClustalO
Clustal is a widely used multiple sequence alignment program that is applicable to both nucleotide and protein sequences. Clustal Omega is the latest addition to the Clustal family, offering a significant increase in scalability over previous versions. It allows hundreds of thousands of sequences to be aligned in just a few hours and can utilize multiple processors when available.
Clustal home page: http://www.clustal.org
If you are using Windows OS, there are no additional configuration steps required, as the ClustalO executable file is included in the UGENE distribution package. Otherwise:
- Install the Clustal program on your system.
- Set the path to the ClustalW executable on the External tools tab of the UGENE Application Settings dialog.
Now you are able to use ClustalO from UGENE.
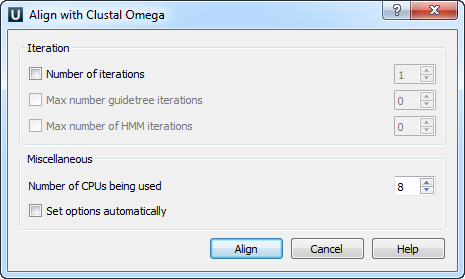
Open a multiple sequence alignment file and select the Align with ClustalO item in the context menu or in the Actions main menu. The Align with ClustalO dialog will appear (see below), where you can adjust the following parameters:
- Number of iterations — number of combined guide tree/HMM iterations.
- Max number of guide tree iterations — maximum number of guide tree iterations.
- Max number of HMM iterations — maximum number of HMM iterations.
- Number of CPUs being used — number of processors to use.
- Set options automatically — set options automatically (might overwrite some of your options).