DNA Flexibility
To search for regions of high DNA helix flexibility in a DNA sequence:
- Open the sequence in the Sequence View.
- Right-click and select Analyze → Find high DNA flexibility regions.
🧬 Note: Only the standard DNA alphabet is supported:
A,C,G,T, andN.
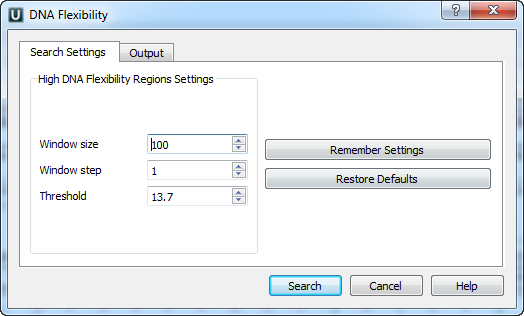
Settings Dialog
The following dialog appears:
Method
The analysis uses a sliding window approach. For each window:
- If 2 or more consecutive windows have average flexibility above the threshold, the region is annotated.
- The average flexibility for a window is calculated as:
Average = (Sum of dinucleotide flexibility angles in the window) / (window size - 1)
Flexibility Angles Table
| Dinucleotide | Angle | Dinucleotide | Angle |
|---|---|---|---|
| AA | 7.6 | CA | 14.6 |
| AC | 10.9 | CC | 7.2 |
| AG | 8.8 | CG | 11.1 |
| AT | 12.5 | CT | 8.8 |
| GA | 8.2 | TA | 25.0 |
| GC | 8.9 | TC | 8.2 |
| GG | 7.2 | TG | 14.6 |
| GT | 10.9 | TT | 7.6 |
Handling Unknown Bases (N)
If the sequence contains an ambiguous base N, the following minimum angle values are used:
- CN, NC, GN, NG, NN → 7.2
- AN, NA, TN, NT → 7.6