Sequence Search Using HMM Profile
The HMM3 search tool reads an HMM profile from a file and searches a sequence for significantly similar sequence matches.
The sequence must be selected in the Project View, or there must be an active Sequence View window opened.
If the selected sequence is nucleic and the profile HMM is built from an amino acid alignment, the sequence will be automatically translated and searched in all possible frames (total of 6).
If a profile HMM is built for nucleotide alignment, the search is performed for both strands (direct and complement).
The HMM3 search accepts the HMMER2 HMM profiles (amino acid only) as a backward compatibility feature. An interesting post about using the HMMER2 models with the HMMER3 is available on Sean Eddy’s blog.
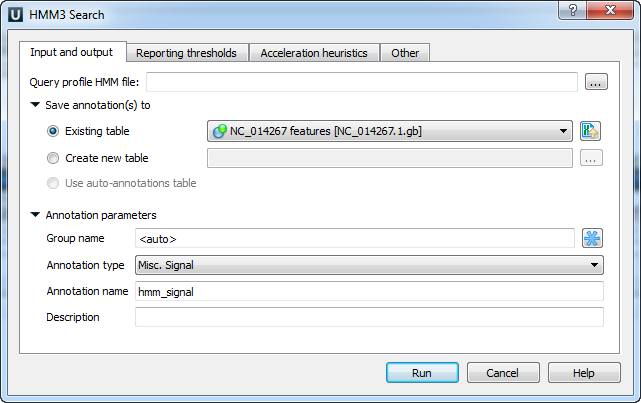
For example, reporting threshold options can be configured using the dialog:
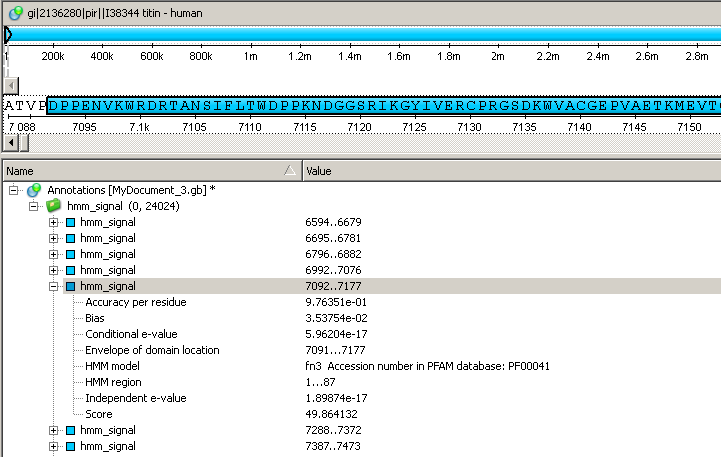
The search results are stored as sequence annotations in the GenBank file format.
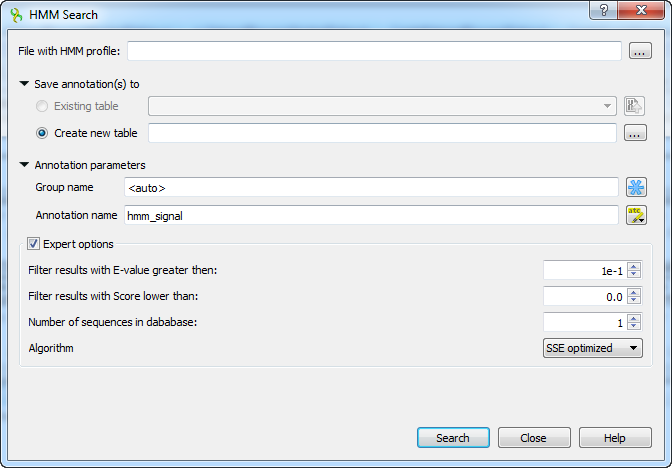
The HMM3 search works only with files that contain a single HMM model.