ORF Marker
In this chapter, you can learn how to search for Open Reading Frames (ORF) within a DNA sequence. The ORFs identified are stored as automatic annotations. This means that if the automatic annotations highlighting has been enabled, ORFs are searched for and highlighted in each sequence opened. Refer to Automatic Annotations Highlighting to learn more.
To open the ORF Marker dialog, select the Analyze ‣ Find ORFs item in the context menu.
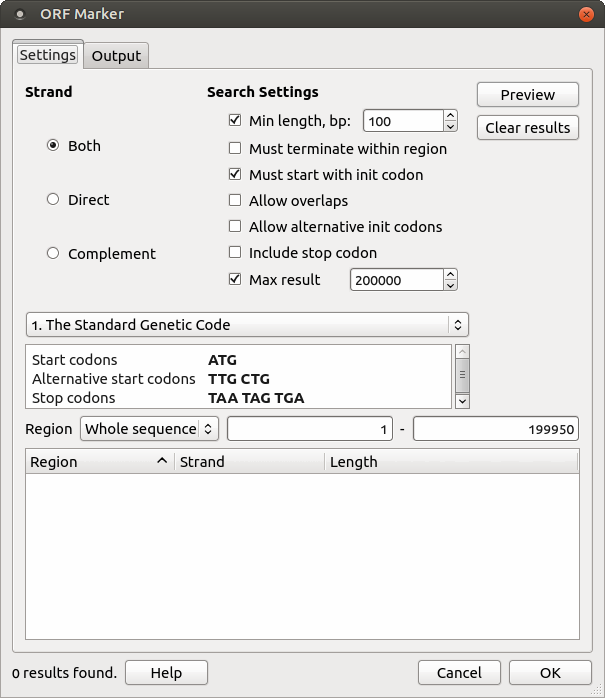
The following Search Settings are available:
- Min length — ORFs with lengths shorter than the Min length value will not be found.
- Must terminate within region — this option ignores boundary ORFs located beyond the search region.
- Must start with init codon — this item switches the ORF Marker algorithm to the mode where any non-stop amino acid code is interpreted as the region start position.
- Allow overlaps — alternative (downstream) initiators, when another start codon is located within a longer ORF, i.e., all possible ORFs will be found, not only the longest ones.
- Allow alternative init codon — this option includes ORFs starting with alternative initiation codons, according to the current translation table.
- Include stop codon — this option includes stop codons in the resulting annotations.
- Max result — this option allows you to define the maximum number of results to be found.
The other available parameters are:
- DNA-to-Amino translation table defines how start, alternative start, and stop codons are encoded.
- Strand — specifies where to search for the ORFs: in the direct strand, in the complement strand, or in both strands.
- Preview — allows you to preview the regions, strands, and lengths of the found ORFs.
- Clear results — becomes available when some results have been found; this clears those results.
To set the saving parameters, go to the Output tab of the dialog.
Here you can modify the annotations saving parameters (Group name, Description, and a file to save the annotation to).
Results:
When the search parameters have been selected and the OK button has been pressed in the dialog, the auto-annotating feature becomes enabled. In the Annotations editor, the ORFs annotations can be found in the Auto-annotations\orf group.
After the search has been completed, you can browse the results, sort them by length, strand, or start position, and save them as annotations to the original sequence in the Genbank format.
For more information about codons, use the codon table. To show or hide the table, use the Ctrl+T shortcut or click the Show codon table toolbar button menu.
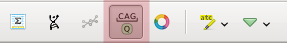
The codon table will appear:
Clicking on a codon name redirects you to Wikipedia, providing a brief description of the corresponding amino acid. The cells of the table are colored according to the classes of amino acids.