PCR Primer Design for DNA Assembly
Overview
The polymerase chain reaction (PCR) uses a pair of custom primers to direct DNA elongation toward each other at opposite ends of the sequence being amplified. These primers are typically between 18 and 24 bases in length and must code for only the specific upstream and downstream sites of the sequence being amplified. A primer that can bind to multiple regions along the DNA will amplify them all, eliminating the purpose of PCR.
A few criteria must be considered when designing a pair of PCR primers. Pairs of primers should have similar melting temperatures since annealing during PCR occurs for both strands simultaneously, and this shared melting temperature must not be too much higher or lower than the reaction’s annealing temperature. A primer with a _T_m (melting temperature) much higher than the reaction’s annealing temperature may mishybridize and extend at an incorrect location along the DNA sequence. A _T_m significantly lower than the annealing temperature may fail to anneal and extend at all.
Additionally, primer sequences need to be chosen to uniquely select for a region of DNA, avoiding the possibility of hybridization to a similar nearby sequence.
There are other parameters to be taken into account - Gibbs free energy, self- and heterodimers availability, etc. All parameters are described below in detail.
UGENE provides the “PCR Primer Design for DNA Assembly” feature only for nucleic sequences with the “Standard DNA” alphabet. To use it in UGENE, open a DNA sequence and go to the “PCR Primer Design for DNA Assembly” tab of the Options Panel:
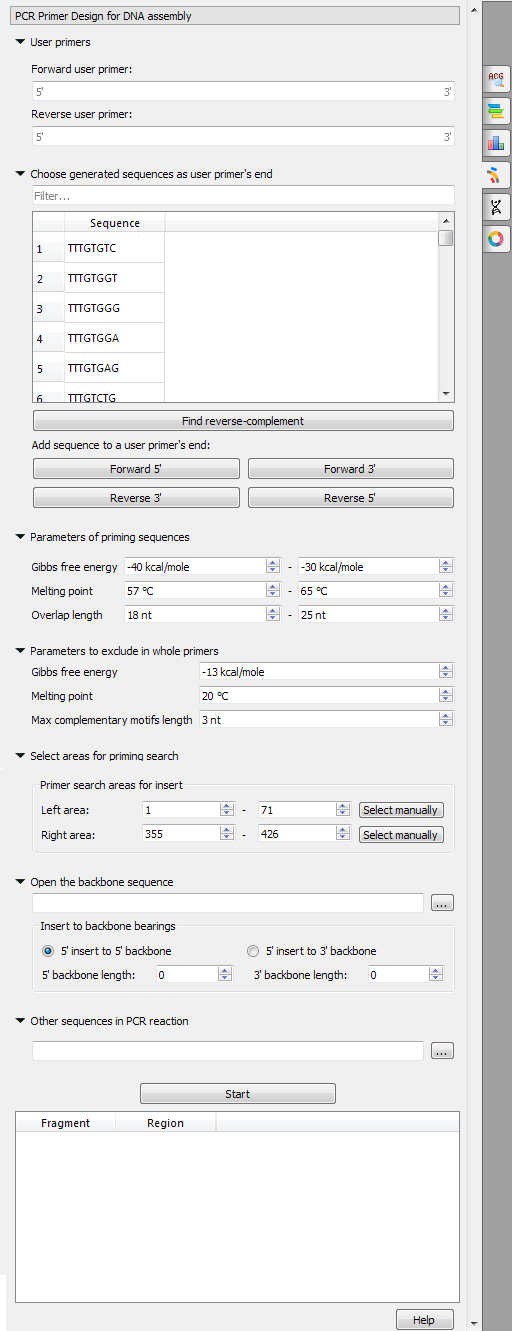
The following parameters are available:
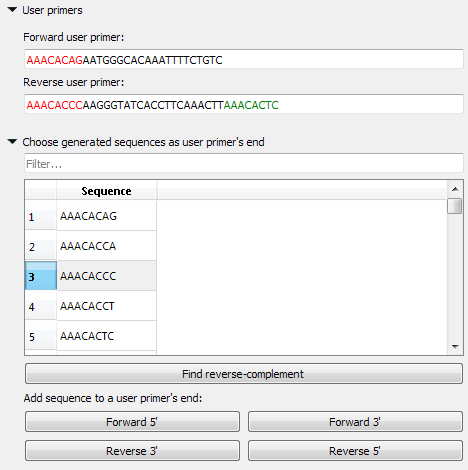
- User primers: The pair of forward and reverse primers can be checked for the absence of unwanted connections (like hairpins, self- and heterodimers).
- Choose generated sequences as user primers’ end: User primers could be extended with 8-base sequences on the 5’ and 3’ ends. These sequences do not form simple unwanted connections (for example, “AAAATTTT” obviously has a hairpin, so such sequences are not included in the list).
- Parameters of priming sequences: The range of melting temperature, Gibbs free energy, and length primers should have.
- Parameters to exclude in whole primers: The minimum melting temperature, maximum Gibbs free energy, and maximum base pair length that may be present in the self- and heterodimers in the considered primer.
- Select areas for priming search: Select areas to search forward and reverse primers in.
- Open the backbone sequence: Choose a sequence to be added to the 5’ or 3’ end of the result primer.
- Other sequences in the PCR reaction: The list of sequences to check for unwanted connections. The unwanted connections should fit the “Parameters to exclude in whole primers”.
Designing Primers
Physical Quantities
The designed primers should have the specified Gibbs free energy, melting temperature, and length. These primers could also have hairpins, self-, and heterodimers, but the mentioned parameters of these found dimers should be limited. See the pictures below:
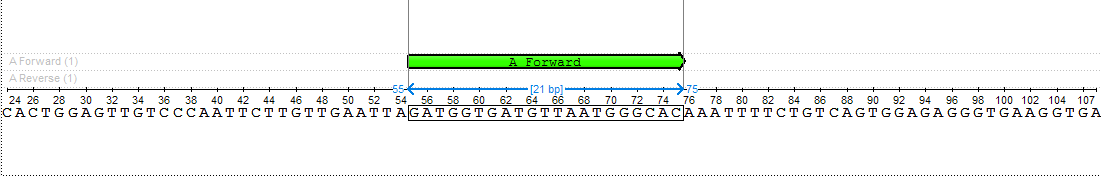
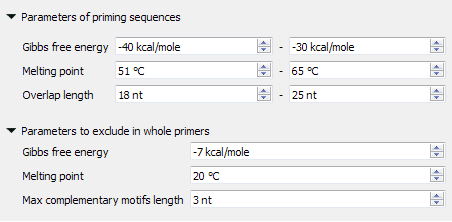
The found primer “GATGGTGATGTTAATGGGCAC” has a Gibbs free energy between -40 and -30 kcal/mol, a melting temperature between 51 and 65 °C, and a length between 18 and 25 nucleotides. Also, there are no hairpins, self-, and heterodimers in this primer, which have Gibbs free energy greater than -7 kcal/mol, melting temperature less than 20 °C, and base pairs length more than 3 nucleotides.
Regions
Forward primers are searched in the left region, while reverse primers are searched in the right region. There is an amplified fragment between them:
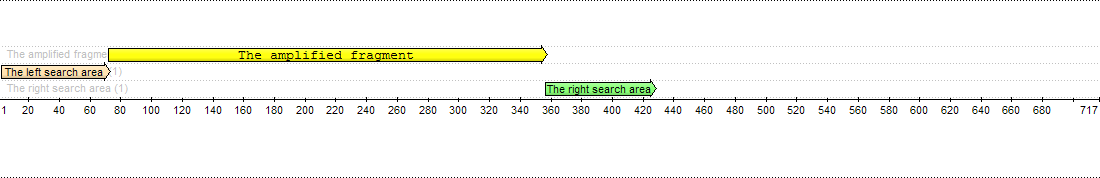
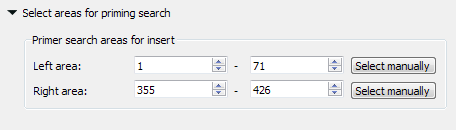
The left search area is from 1 to 71 bases, and the right search area is from 355 to 426 bases. The amplified fragment is between them.
Backbone Sequence
The backbone sequence is the sequence added to the 5’ end of the primer for the following assembly of the PCR product with the backbone molecule. This parameter is optional. You may set the backbone sequence, the sequence end you want to add the backbone to, and the length of the sequence from the 5’ and 3’ ends to check if these sequences have unwanted hairpins, self-, and heterodimers. The pictures below show the differences between “5’ insert to 5’ backbone” and “5’ insert to 3’ backbone” (“insert” in this case is the same as “primer”).
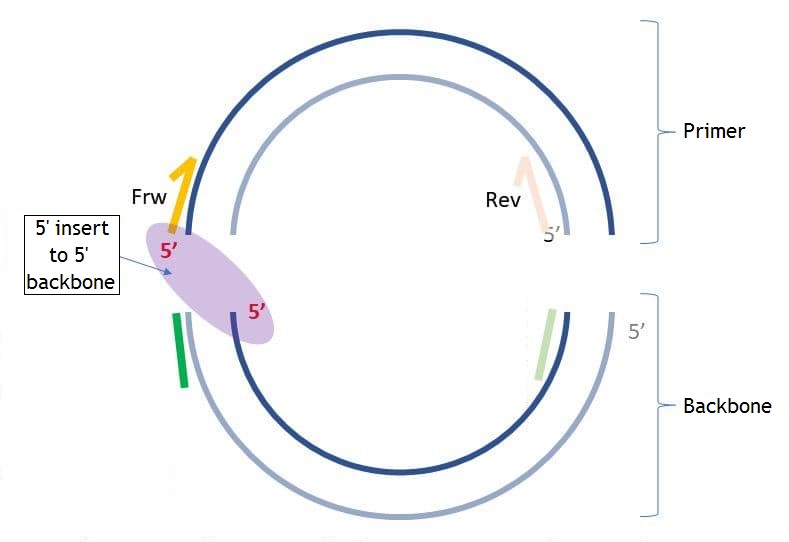
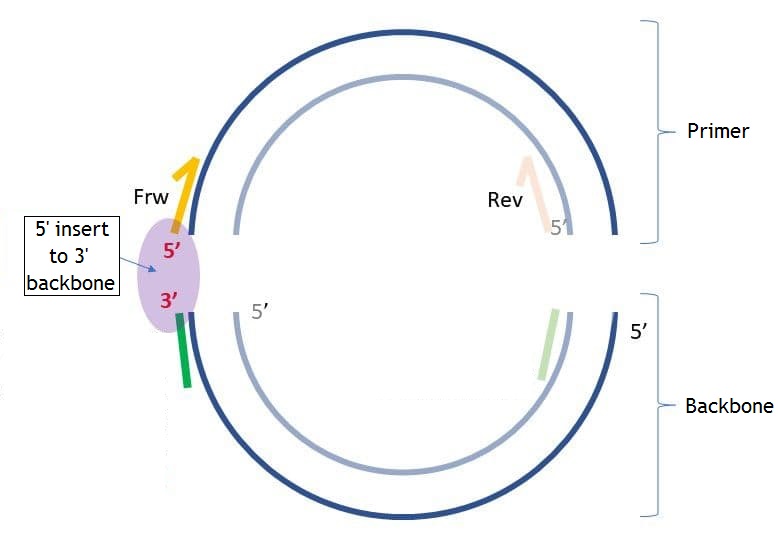
If you set “5’ backbone length,” then the number of bases you set from the 5’ end of the backbone sequence will be checked for unwanted connections. The same applies to “3’ backbone length,” but from the 3’ end. If the checked end has hairpins, self-, and homodimers, you will see the following dialog:
Other Sequences in PCR Reactions
Sequences that are supposed to be in the reaction mixture. It is important that primers do not have unwanted connections to these sequences, so it will be checked that there are no hairpins, self-, or heterodimers between the resulting primer and any sequence from the “Other sequences in PCR reaction” file.
Result
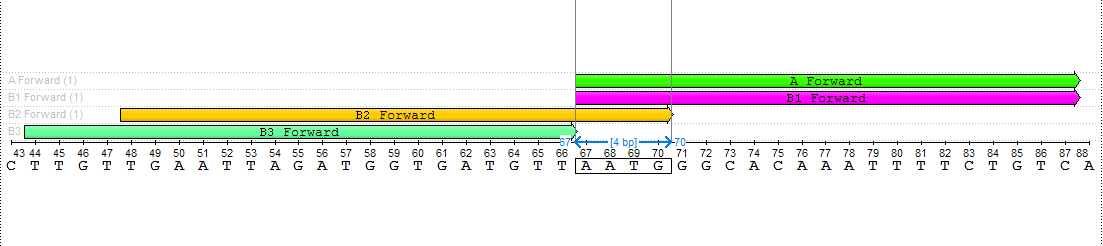
The search result is from 1 to 4 pairs of primers. The results should be as close as possible to the amplified fragment. This means that the searching process starts from the edge of the amplified fragment in the 5’ direction on the direct strand for the forward primer and in the 3’ direction on the reverse-complementary strand for the reverse primer. The results are defined with special names:
- A: The pair of primers closest to the amplified fragment, corresponding to the entire set of parameters.
- B1: The pair of primers closest to the amplified fragment, corresponding to the entire set of parameters if B2 and/or B3 exist.
- B2: The pair of primers corresponding to the entire set of parameters, which has a 4-nucleotide intersection with B1.
- B3: The pair of primers corresponding to the entire set of parameters, which starts immediately when B1 ends.
You may also see results in the result table. A single click on the result in the result table selects the corresponding annotation, while double-clicking exports the resulting primer to a separate file (with the backbone sequence, which will be added during the exporting process).
User Primer
You may check the validity of the pair of primers by typing this pair into the corresponding fields. You may also add your primers’ 5’ and/or 3’ 8-base endings, which do not form simple unwanted connections.