Primer3
The Primer3 plugin is a port of the Primer3 tool, v. 2.6.1. It is intended to pick primers from a DNA sequence.
Usage
To use Primer3, open a DNA sequence and select the Analyze ‣ Primer3 context menu item. The dialog will appear:
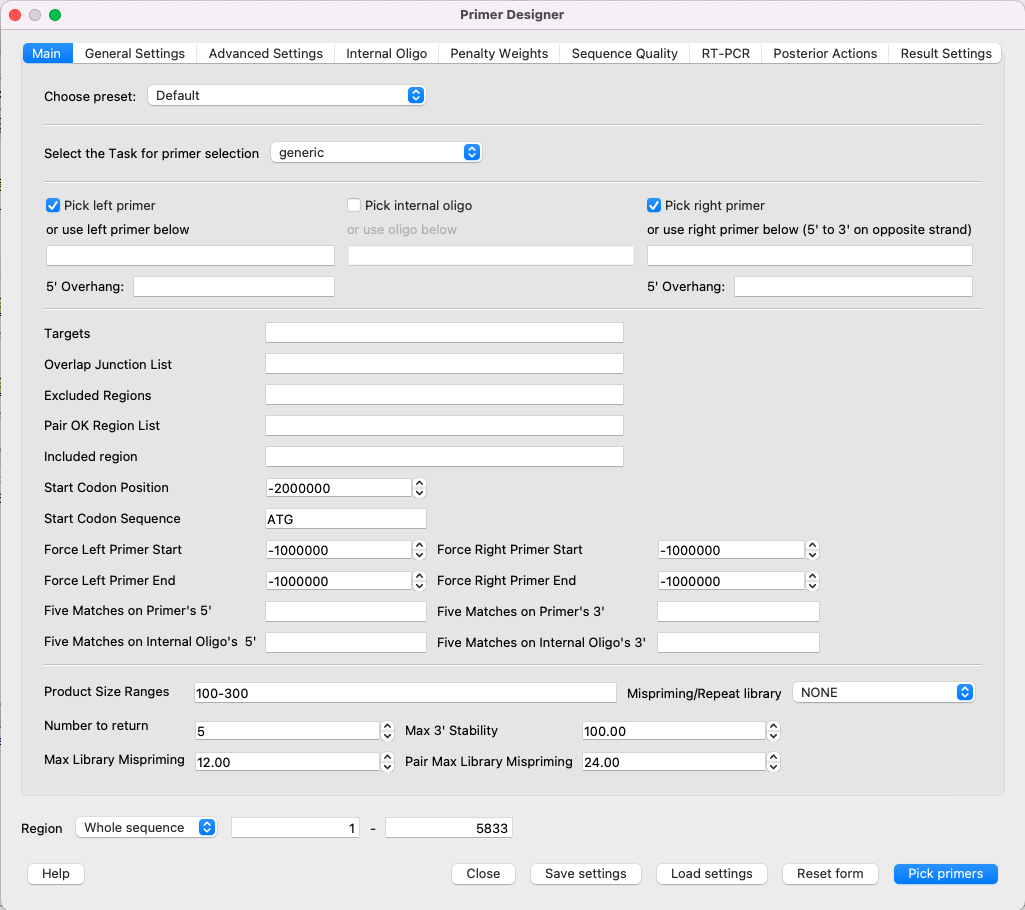
All available parameters are the same as the original Primer3.
NOTE: The “Primer Designer” dialog has a detailed description of each parameter. Just move the mouse cursor to the parameter you’re interested in (or its name), and the tooltip appears. Therefore, there is no detailed description of each parameter in this file.
Report
Once the calculation is finished, a notification appears in the bottom right corner. Click on it to open the report file.
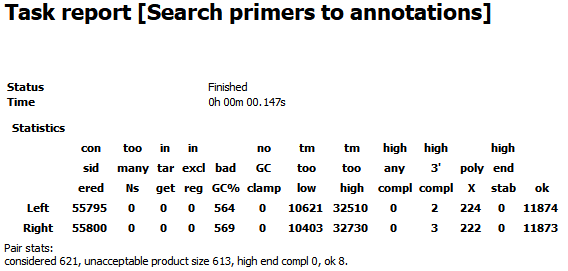
The report contains information about how many primers were considered during the search process and how many of them were filtered for various reasons.
Results
All found primers will be available as annotations and marked on the target sequence:
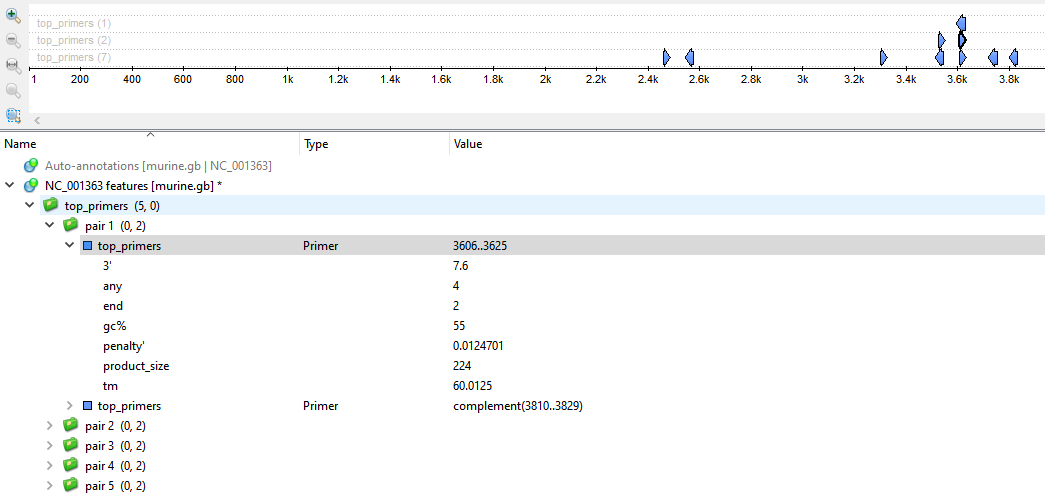
The result annotations could have a set of the following qualifiers:
- 3’ - Free Gibbs energy of 5 bases on the 3’ end.
- any - The tendency of this primer to bind its reverse-complement chain. This value is calculated as follows: the primer aligns on its reverse-complement representation; each match counts +1 point, each mismatch -1 point, each gap -2 points, each N -0.25 points.
- end - Similar to “any,” but only concerning 5 bases on the 3’ end.
- gc - GC content.
- penalty - Penalty weight (see this article for details).
- hairpin - A hairpin loop melting temperature (if this kind of loop exists in the primer).
- product_size - Size of the product.
- tm - Primer melting temperature.
There is one additional feature available that is not originally part of the Primer3 tool. It allows users to design primers for RT-PCR experiments by choosing which exons/introns to span with the primer product. This feature is described in detail below. When you select the parameters, you can save and load settings with the help of the corresponding buttons in the right corner of the dialog.