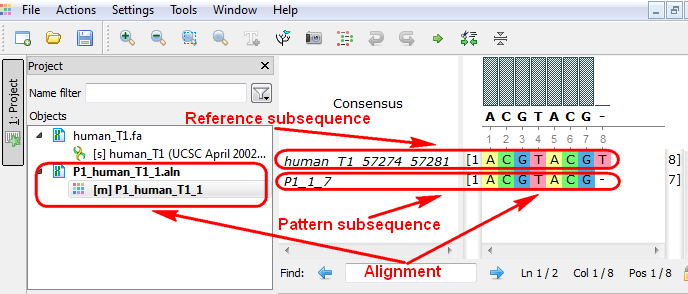
Smith-Waterman Search
The Smith-Waterman Search plugin adds a complete implementation of the Smith-Waterman algorithm to UGENE.
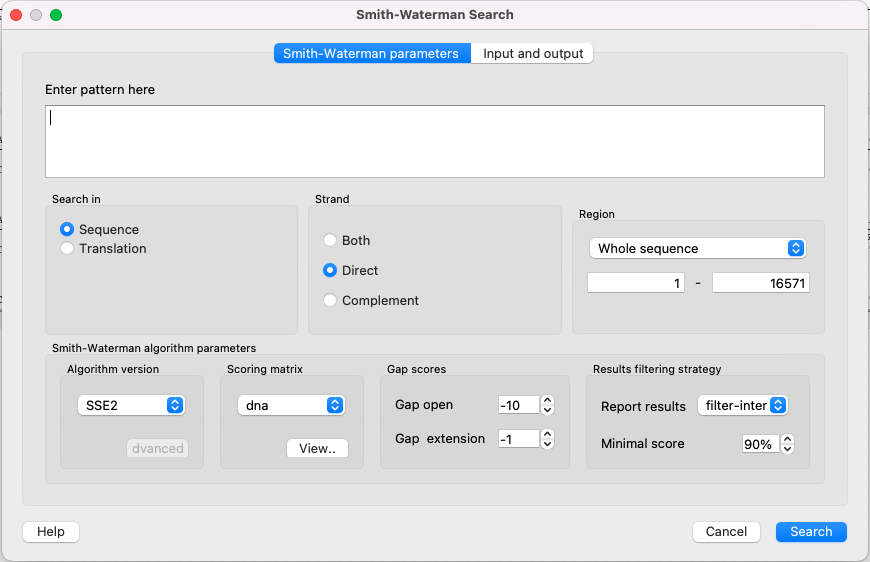
To use the plugin, open a nucleotide or protein sequence in the Sequence View and select the Analyze ‣ Find pattern [Smith-Waterman] item in the context menu. The Smith-Waterman Search dialog appears:
Algorithm
This search algorithm has a significant advantage in calculation time over the usual Search in sequence algorithm for large sequences. This happens because the “Search in sequence” algorithm represents a suffix algorithm - one sequence moves along the other, and areas opposite each other are compared. Using this approach, the computation time increases in direct proportion to the length of the sequence. The Smith-Waterman algorithm uses a completely different approach - searching using a certain kind of matrix.
The algorithm works in the following way:
A matrix is created where the sequence in which the search is carried out is located vertically, and the searched sequence is located horizontally. Fill the adjacent row and column with 0. Example: The sequence in which the search is carried out - ACGCGAT. The searched sequence - CGCG.
D(i,j) C G C G 0 0 0 0 0 A 0 C 0 G 0 C 0 G 0 A 0 T 0Start filling the matrix from the top-left corner, moving from left to right and from top to bottom, using the following formula:
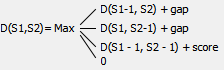 Where:
D(S1, S2) - current cell,
D(S1-1, S2) - the cell on the left,
D(S1, S2-1) - the cell above,
D(S1-1, S2-1) - the one on the top and left,
gap - the Gap open penalty if we encounter a gap for the first time and the Gap extension penalty if we encounter a gap the second time or more.
score - the value from the Scoring matrix table.
The following example uses nucl Scoring matrix (+5 for match, -4 for mismatch) and -1 for Gap open and Gap extension penalties:
Where:
D(S1, S2) - current cell,
D(S1-1, S2) - the cell on the left,
D(S1, S2-1) - the cell above,
D(S1-1, S2-1) - the one on the top and left,
gap - the Gap open penalty if we encounter a gap for the first time and the Gap extension penalty if we encounter a gap the second time or more.
score - the value from the Scoring matrix table.
The following example uses nucl Scoring matrix (+5 for match, -4 for mismatch) and -1 for Gap open and Gap extension penalties: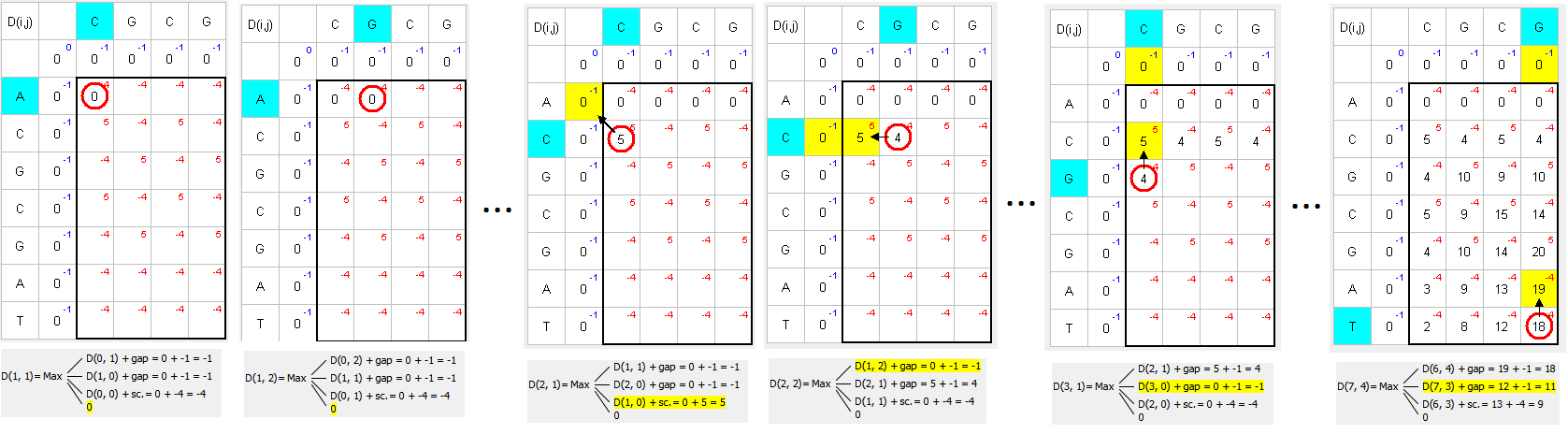
Find the largest number and move to the top, left, or top-left to the next larger number:
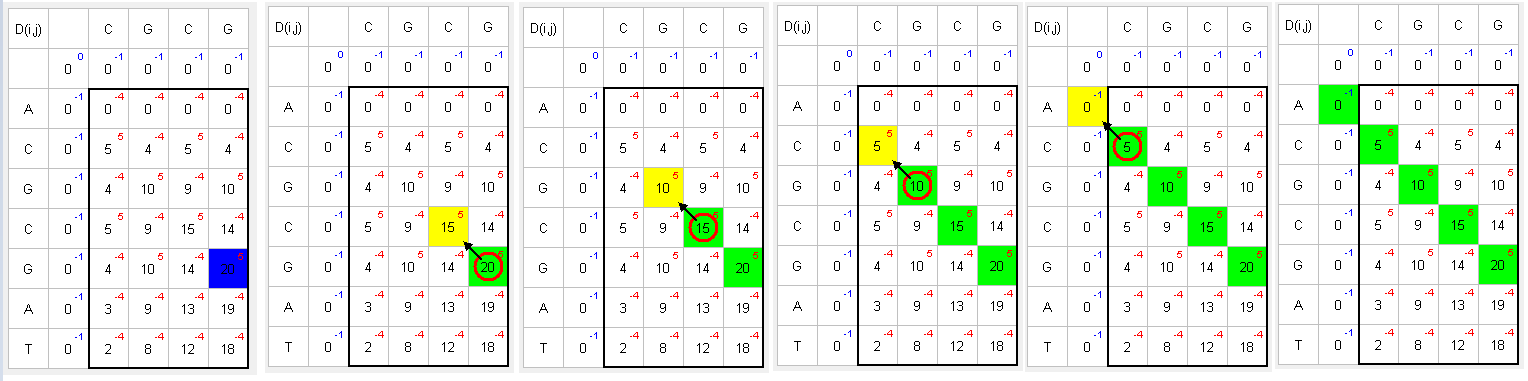 Matching green symbols indicate the desired sequence intersection.
Matching green symbols indicate the desired sequence intersection.
Parameters
First of all, you need to specify the pattern to search for. The rest of the parameters are optional:
Search in — select either to search in the sequence or in its amino acid translation.
Strand — select the strand to search in: direct, reverse-complementary, or both strands.
Region — specifies the region of the sequence that will be used to search for the pattern. By default, if a subsequence has been selected when the dialog has been opened, then the selected subsequence is searched for the pattern. Otherwise, the whole sequence is used. You can also input a custom range.
Algorithm version — version of the algorithm implementation. All versions produce the same result - only the inner technology is used differently:
- Classic 2 - classical implementation of the algorithm. Works everywhere.
- SSE2 - the algorithm, which works much faster, but requires you to have an SSE2 processor.
Scoring matrix — can be chosen from a variety of matrices supplied with UGENE. To view a selected matrix, click the View button.
Gap open — penalty for opening a gap.
Gap extension — penalty for extending a gap.
Report results — simple heuristic which allows filtering of intersected hits. If it is set to none, the algorithm may report a large set of almost identical results in the same region.
Minimal score — another simple heuristic which measures sequence similarity. It is more convenient than using some abstract scores. If set to 100%, the algorithm will search for an exact substring match.
Input and output
The results of the search are saved as annotations or as multiple alignments. To set the saving parameters, go to the Input and output tab of the dialog.
If you want to save the results as annotations, input the annotations saving parameters (Annotation name, Group name, Annotation type, Description and a file to save the annotation to). You can also add a qualifier with corresponding pattern subsequences to result annotations. Check the corresponding checkbox for this.
If you want to save the results as multiple alignments, select the following parameters:
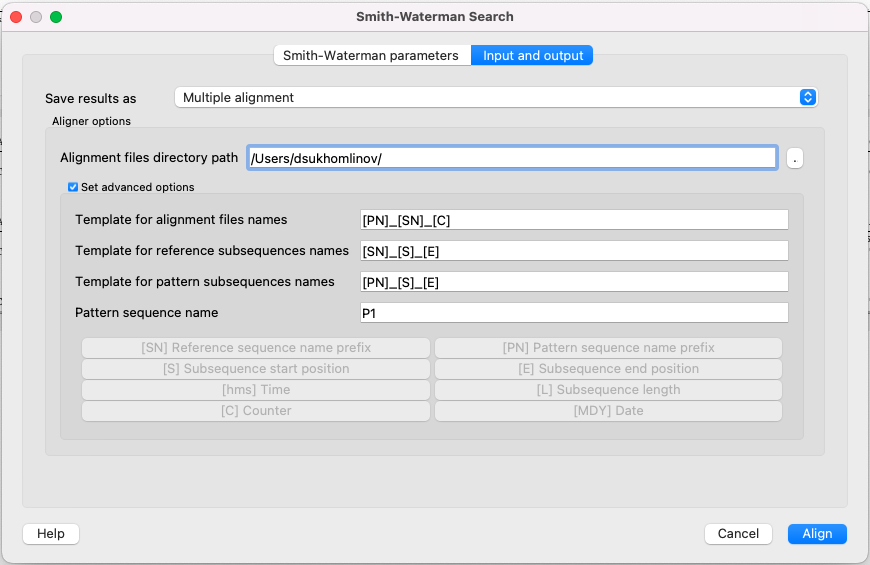
Here you can select a file to save the alignment to (Alignment files directory path parameter).
Using the Set advanced options checkbox, you can select the saving options.
You can set different templates for file names: create your own or create using the following: [E] — adds a subsequence end position, [hms] — adds a time, [MDY] — adds a date, [S] — adds a subsequence start position, [L] — adds a subsequence length, [SN] — adds a reference sequence name prefix, [PN] — adds a pattern sequence name prefix, [C] — adds a counter.
You can create templates for alignment file names, reference subsequence names, pattern subsequence names, and for the pattern sequence name: