Building Index for UGENE Genome Aligner
You can build an index to optimize short reads alignment using UGENE Genome Aligner. To open the Build Index dialog, select the Tools ‣ NGS data analysis ‣ Build index for reads mapping item in the main menu. Set the value of the Map short reads method parameter to UGENE Genome Aligner.
The dialog looks as follows:
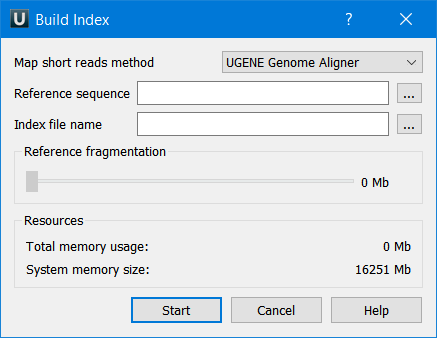
The parameters are the following:
- Reference sequence — DNA sequence to which short reads will be aligned. This parameter is required.
- Index file name — file to save the index to. This parameter is required.
- Reference fragmentation — this parameter influences the number of parts into which the reference will be divided. It is better to make it larger, but it increases the amount of memory used during the alignment.
- Total memory usage — shows the total memory usage.
- System memory size — shows the total system memory size.