Weight Matrix
The Weight Matrix plugin is a tool for solving the problem of sequence annotation. Much like the SITECON, the main use case of the plugin is the recognition of potential transcription factor binding sites based on data about conservative conformational and physicochemical properties revealed through binding site set analysis.
The Weight Matrix contains numerous position frequency matrices (PFMs) and position weight matrices (PWMs, also known as position-specific score matrices — PSSMs). These matrices come from two widely-known open archives: JASPAR, which contains frequency matrices, and UniPROBE containing weight matrices.
Additionally, the Weight Matrix plugin provides a tool for creating specific position frequency and weight matrices from an existing alignment or from a file with several sequences. The created matrix can be used as a profile for the search, just like the JASPAR and UNIPROBE ones.
To search for transcription factor binding sites in a DNA sequence, select the Analyze ‣ Search TFBS with matrices context menu item. The Weight matrix search dialog will appear:
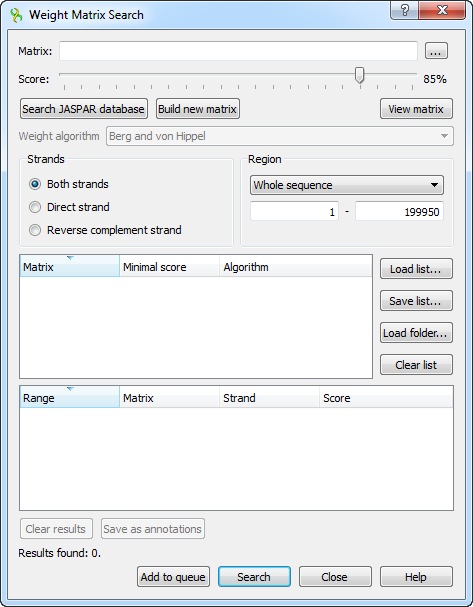
In the search dialog, you must specify a file with PWM or PFM. You can do so by pressing the browse button and selecting the file.
You can also use the special interface to choose a JASPAR matrix by pressing the Search JASPAR database button.
An alternative way to specify the position weight/frequency matrix is to create a specific one from an alignment or a file with several sequences using the build a new matrix tool.
After the profile (the matrix) is loaded, you can adjust the threshold value. The threshold sets the minimal identity score for a result to pass. The higher the result score, the more it is homologously related to the aligned region. By changing the threshold, you can filter low-scoring results.
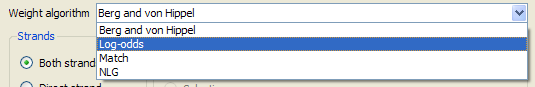
If the loaded matrix is a position frequency matrix, you must also specify the algorithm to build the corresponding position weight matrix, which will represent the transcription factor. There are four algorithms available.
You can also add a selected matrix with the specified Minimal score and the Algorithm to the matrices list. To do this, select the matrix and other options and press the Add to queue button. The plugin will search with all matrices specified in the list.
You can use the Save list button to export the list of matrices to a *.csv file. Later the list can be loaded from the file using the Load list button.
The remaining options are standard sequence search options: the strand and the sequence region where you want to search for matches.
After specifying the necessary options, press the Search button. The found results will appear in the dialog table. The corresponding results identity scores are in the Score column.
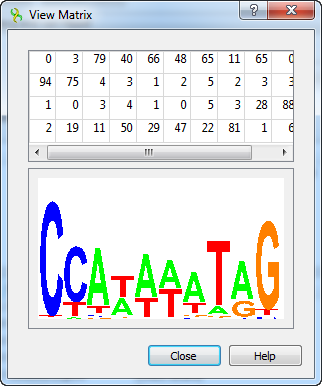
You can also view the matrix by using the View matrix button:
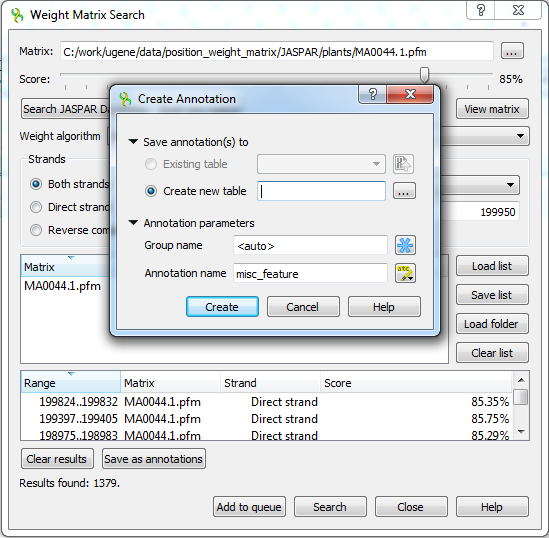
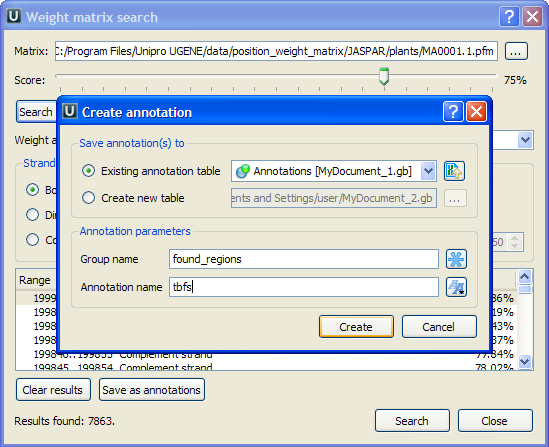
The regions found by the weight matrix algorithm can be saved as annotations to the DNA sequence in Genbank format by pressing the Save as annotations button.
After saving, the file with resulting annotations will be automatically added to the current project, and the annotations will be added to the original sequence.
Note that in the case of selecting a JASPAR or UNIPROBE matrix, the resulting annotations will contain the given matrix properties.
See also: