Mapping Reads to Reference
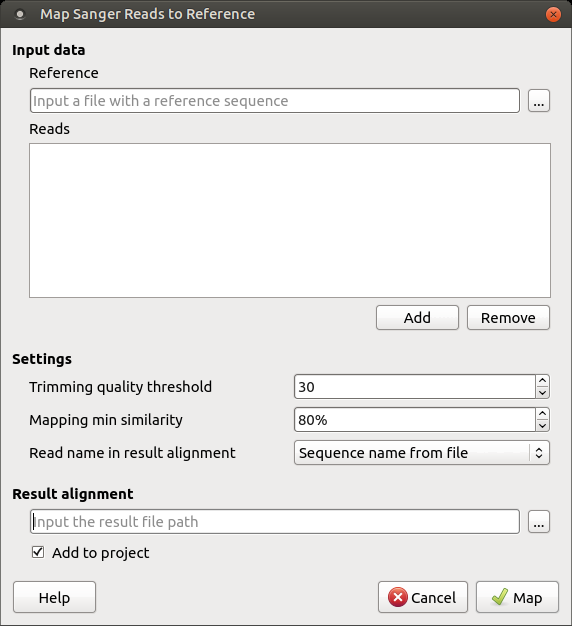
To map Sanger sequencing reads to a reference, use the Tools–> Sanger data analysis–> Map reads to reference main menu item. The following dialog will appear:

The following inputs are required:
Reference sequence — a file with a single DNA sequence from any supported file format (e.g., FASTA or GenBank). This parameter is mandatory.
Reads — a set of files in *.ab1 or *.scf format with the Sanger sequencing data to be mapped to the reference sequence. At least one read must be added. The read orientation (forward or reverse) will be automatically detected during the mapping.
During the execution of the mapping task, these data are processed as follows: Firstly, to enhance further mapping sensitivity, the low-quality ends of the reads are trimmed off. Then, the reads are mapped using UGENE’s original method, which works in two steps: rough mapping of a read using BLAST+, followed by enhancement of the read alignment with the Smith-Waterman algorithm. After that, the percentage similarity of the two sequences is calculated, which is the number of all edit operations (i.e., insertions, deletions, and substitutions) required to transform a read sequence into the corresponding region of the reference sequence. Reads with low similarity to the reference sequence are filtered out.
The following parameters of the mapping task can be configured:
Trimming quality threshold — all bases at the ends of the reads with quality lower than the specified value are trimmed. To skip trimming, set this value to zero.
Mapping min similarity — all reads mapped to the reference with a lower percentage similarity than the specified value are filtered out.
Read name in result alignment — reads in the resulting alignment can be named either by the names of the sequences in the input files or by the input file names. Set this value to “File name”. For example, if the sequences in the input *.ab1 files have the same name, this will help distinguish the reads in the resulting alignment.
The resulting alignment is stored in a native UGENEDB format. You can set up the file location and name in the Result alignment field. Note that it is also possible to export data to standard alignment formats without chromatograms such as FASTA, ClustalW, etc.
To initiate the execution of the mapping task, click the Map button in the dialog. Note that when the task is finished, task statistics can be found in a report, available by clicking the corresponding notification: