Sequence View Components
The Sequence View is one of the major Object Views in UGENE, designed to visualize and edit DNA, RNA, or protein sequences alongside their properties such as annotations, chromatograms, 3D models, statistical data, etc.
For each file, UGENE analyzes the file content and automatically opens the most appropriate view.
To activate the Sequence View, open any file containing at least one sequence. For example, you can use the $UGENE/data/samples/EMBL/AF177870.emb file provided with UGENE.
After opening the file in UGENE, the Sequence View window appears:
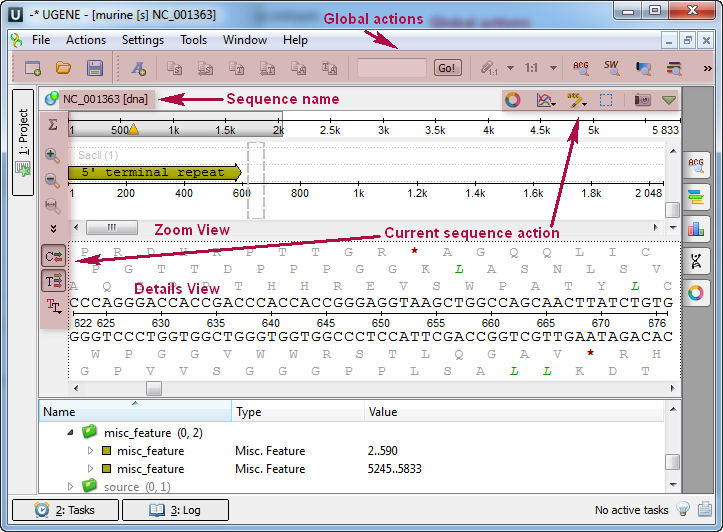
Once the view is opened, you will see a set of new buttons in the toolbar area. The actions provided by these buttons are available for all sequences opened in the view. In the picture below, these buttons are indicated by the “Global actions” arrow.
Below the toolbar, there is an area for one or several sequences. For each sequence, a smaller toolbar with actions for the sequence is available, along with the following areas:
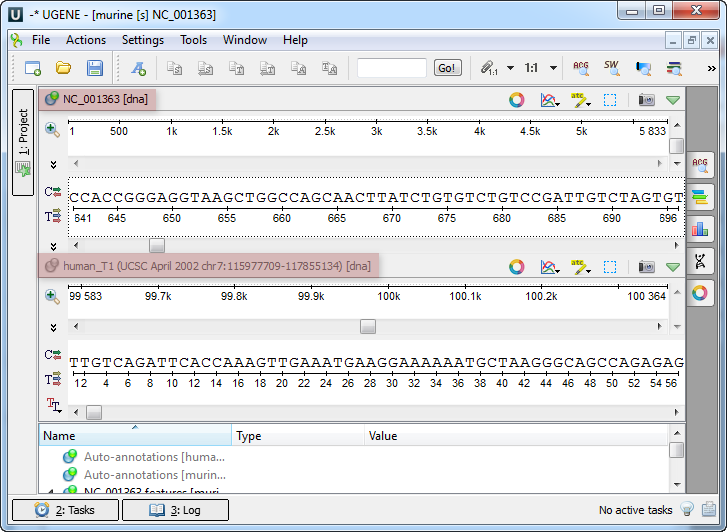
An example of the Sequence View with several sequences:
You can change the focus by clicking on the corresponding sequence area. All sequences that are not in focus will have their sequence name and icon disabled.
The bottom area of the Sequence View is the Annotations Editor. It contains a tree-like structure of all annotations available for all sequences shown in the Sequence View and can be used to perform various actions on annotations: create a new annotation, modify an existing one, group, sort, etc.