Extract Consensus as Sequence
For each input multiple alignment, the workflow calculates the consensus and saves it to a FASTA file, named according to the input alignment’s name.
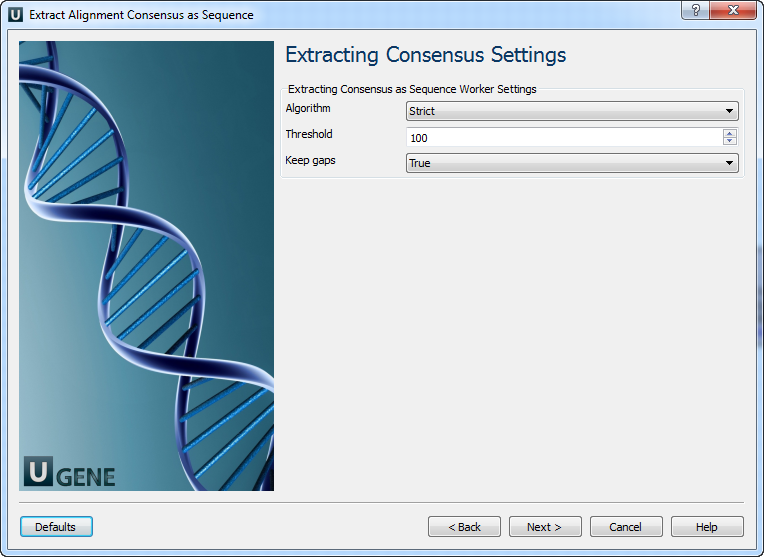
The “strict” algorithm, with the “threshold” parameter set to “100%”, is used by default to calculate the consensus. This means that the consensus will only contain characters that are the same in all sequences of the alignment. Decreasing the threshold will result in considering only the specified percentage of the sequences. For example, if the threshold is 80% and 82% of the sequences have “A” at a certain column position, the consensus will also contain “A” at this position.
Alternatively, you may select another algorithm to calculate the consensus. The algorithm proposed by Victor Levitsky uses the extended DNA alphabet. The greater the threshold value selected for this algorithm, the more rare characters are taken into account. The specified value must be between 50% and 100%.
Finally, there is a “Keep gaps” parameter that specifies whether the resulting sequence must contain gaps or whether they should be skipped. By default, gaps are kept in the resulting consensus sequence.
How to Use This Sample
If you haven’t used the workflow samples in UGENE before, refer to the How to Use Sample Workflows section of the documentation.
Workflow Sample Location
The sample “Extract Consensus as Sequence” can be found in the “Alignment” section of the Workflow Designer samples.
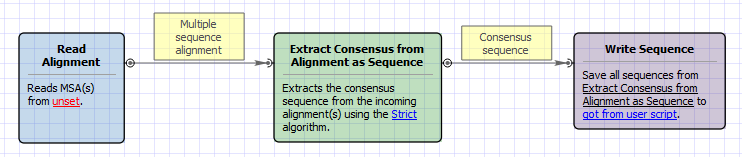
Workflow Image
The opened workflow looks as follows:
Workflow Wizard
The wizard has 3 pages:
1. Input Multiple Alignments
On this page, you must input one or more multiple alignment files.
2. Extracting Consensus Settings
On this page, you can configure consensus extraction parameters.
| Parameter | Description |
|---|---|
| Algorithm | The algorithm used to extract the consensus |
| Threshold | The minimum percentage to include a character |
| Keep gaps | Whether to preserve gaps in the consensus sequence |
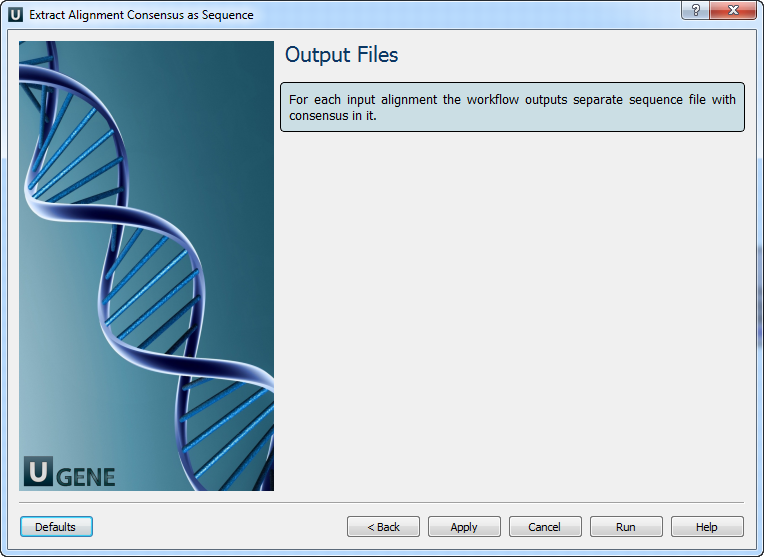
3. Output Files
For each input alignment, the workflow produces a separate FASTA file with the consensus sequence.