Convert seq-qual Pair to FASTQ
This workflow allows you to add PHRED quality scores to a sequence and save the result in FASTQ format. For example, you can read a FASTA file, import PHRED quality values from a corresponding qualities file, and export the result to FASTQ.
How to Use This Sample
If you haven’t used the workflow samples in UGENE before, see the “How to Use Sample Workflows” section of the documentation.
Workflow Sample Location
The workflow sample “Convert seq/qual Pair to FASTQ” is available in the “Conversions” section of the Workflow Designer samples.
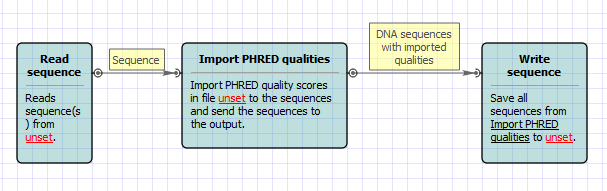
Workflow Image
The workflow looks as follows:
Workflow Wizard
The wizard includes two pages:
1. Input Sequence(s)
On this page, you must input the sequence(s).
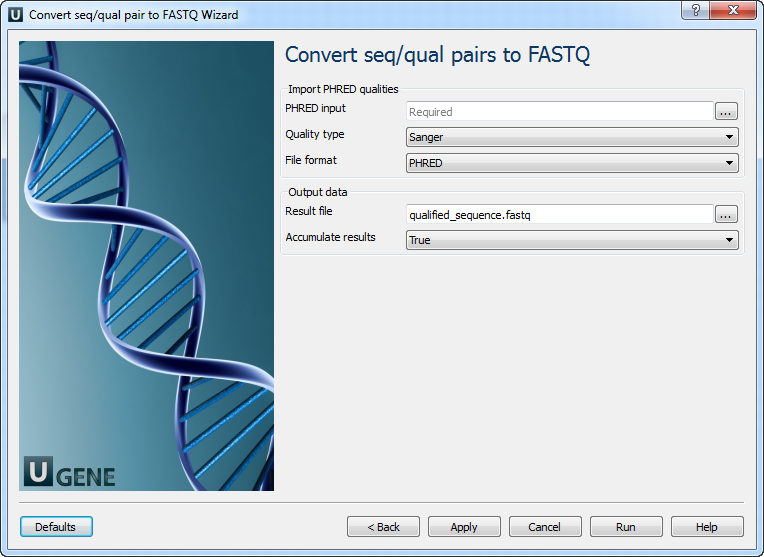
2. Convert “seq/qual” Pair to FASTQ
On this page, you can modify conversion and output settings.
| Parameter | Description |
|---|---|
| PHRED input | Path to file with PHRED quality scores. |
| Quality type | Choose the method to encode quality scores. |
| File format | Quality values format: specialized FASTA-like PHRED or FASTQ-style. |
| Result file | Output file location. If set, it overrides slot “Location” in port. |
| Accumulate results | Accumulate data in one file or generate separate files with an incremental suffix for each input file. |