Convert Sequence to Genbank
This workflow converts sequence file(s) of any format (including PDB, alignments, etc.) to GenBank documents. If the source format supports annotations, they are saved as feature tables in the target file. Sequence meta-information (accessions, etc.) is preserved as well.
How to Use This Sample
If you haven’t used the workflow samples in UGENE before, refer to the “How to Use Sample Workflows” section of the documentation.
Workflow Sample Location
The workflow sample “Convert Sequence to Genbank” is located in the “Conversions” section of the Workflow Designer samples.
Workflow Image
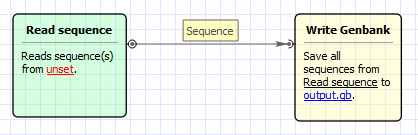
The workflow looks as follows:
Workflow Wizard
The wizard includes 2 pages:
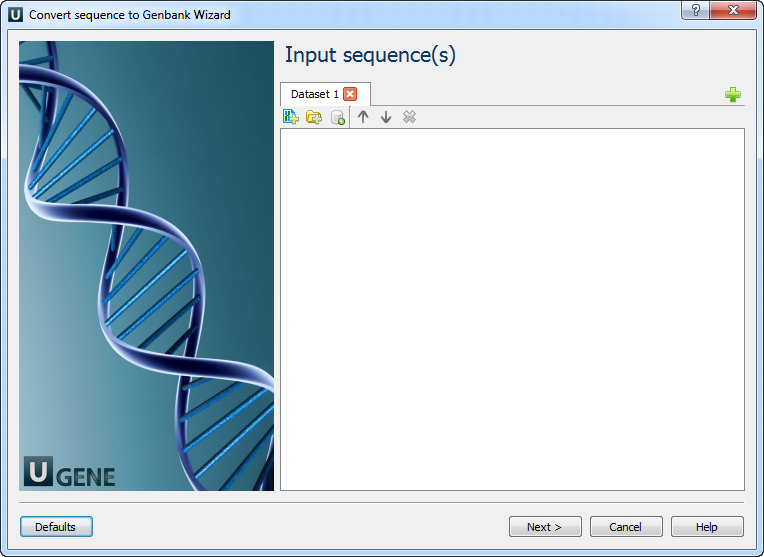
1. Input Sequence(s)
On this page, you must input the sequence(s).
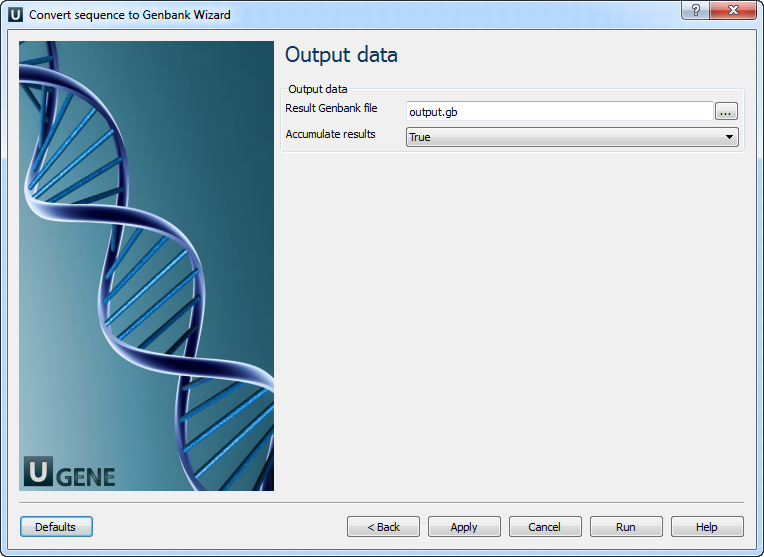
2. Output Data
Modify output file settings.
| Parameter | Description |
|---|---|
| Result Genbank file | Location of the output data file. If this attribute is set, it overrides the “Location” slot of the output port. |
| Accumulate results | Accumulate all input data in one file, or create separate files for each input. The latter adds a numeric suffix per file. |