Build HMM from Alignment and Test It
This workflow builds a new profile HMM from input alignment, calibrates the HMM, and saves it to a file. It then runs a test HMM search over a sample sequence and saves the test results to a Genbank file.
How to Use This Sample
If you haven’t used the workflow samples in UGENE before, please refer to the “How to Use Sample Workflows” section of the documentation.
Workflow Sample Location
The workflow sample “Build HMM from Alignment and Test It” can be found in the “HMMER” section of the Workflow Designer samples.
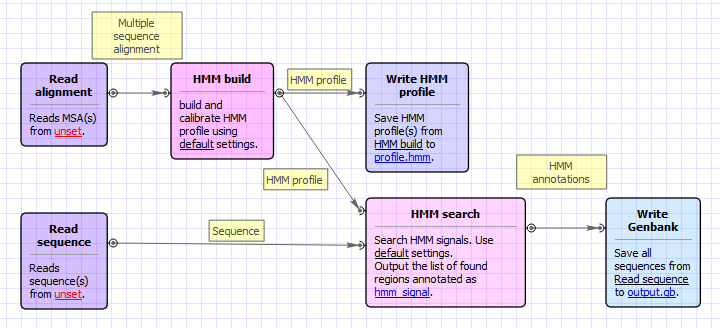
Workflow Image
The workflow appears as follows:
Workflow Wizard
The wizard has 4 pages.
1. Input MSA(s)
On this page, you must input MSA(s).
2. Input Sequence(s)
On this page, you must input sequences.
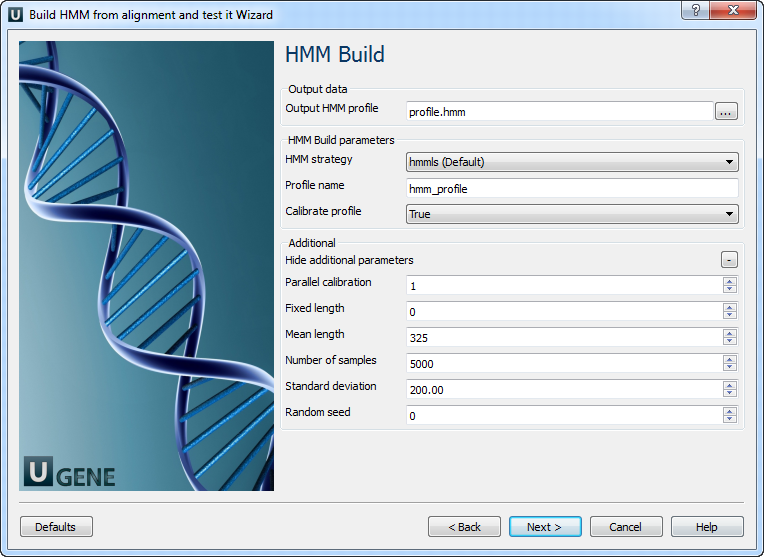
3. HMM Build
On this page, you can modify HMM build parameters.
The following parameters are available:
| Parameter | Description |
|---|---|
| Output HMM profile | Location of the output data file. If this attribute is set, the slot “Location” in port will not be used. |
| HMM strategy | Specifies the kind of alignments you want to allow. |
| Profile name | Descriptive name of the HMM profile. |
| Calibrate profile | Enables/disables optional profile calibration. Calibration increases the sensitivity of the database search. |
| Parallel calibration | Number of parallel threads for calibration. |
| Fixed length | Fixes the length of generated random sequences. The default is to use various lengths from a Gaussian distribution. |
| Mean length | Mean length of synthetic sequences. Default: 325. |
| Number of samples | Number of synthetic sequences. Default: 5000. Too few may fail EVD fitting; more improves accuracy. |
| Standard deviation | Standard deviation of sequence lengths. Default: 200. Gaussian is left-truncated (no zero-lengths). |
| Random seed | Random seed. Use a positive integer for reproducible results. By default, the current time is used, so results vary between runs. |
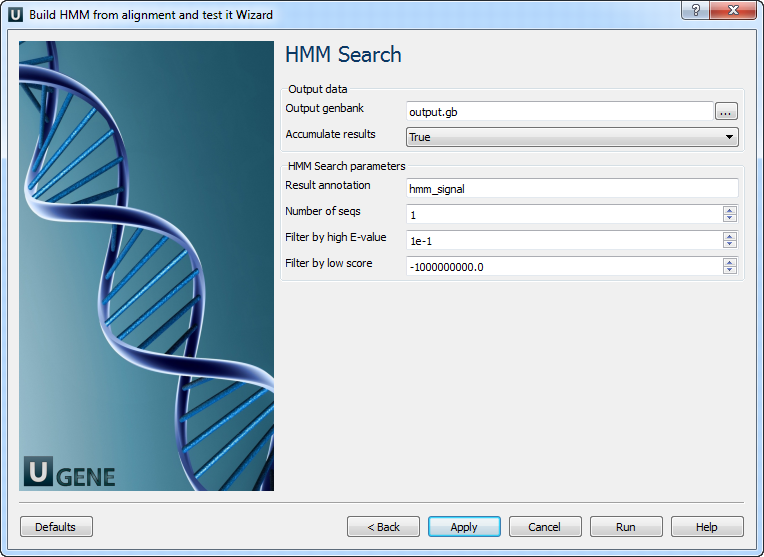
4. HMM Search
On this page, you can modify HMM search and output parameters.
The following parameters are available:
| Parameter | Description |
|---|---|
| Output Genbank | Location of the output data file. If set, the port “Location” is not used. |
| Accumulate results | Combine all results into one file or create separate files (with numeric suffixes). |
| Result annotation | Name of the result annotations. |
| Number of seqs | Calculate E-values as if the sequence DB had this many entries. |
| Filter by high E-value | Use an E-value threshold to exclude low-probability hits. |
| Filter by low score | An alternative to E-value filtering — excludes low-score hits. |