Search Sequences with Profile HMM
This workflow reads an HMM from a file and searches input sequences for significantly similar matches, saving the detected signals to a file. You can specify multiple input files for both the HMM and sequences; the workflow will process the Cartesian product of the inputs. That is, each sequence will be searched with all specified HMMs in turn.
How to Use This Sample
If you haven’t used the workflow samples in UGENE before, consult the “How to Use Sample Workflows” section of the documentation.
Workflow Sample Location
The workflow sample “Search Sequences with Profile HMM” can be found in the “HMMER” section of the Workflow Designer samples.
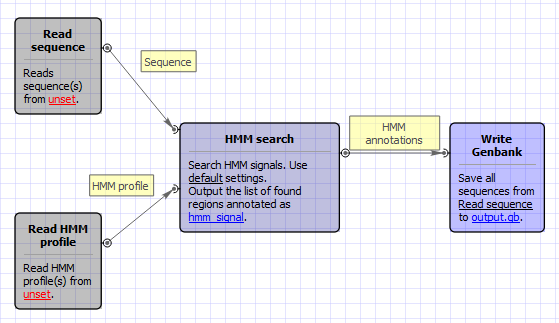
Workflow Image
The workflow appears as follows:
Workflow Wizard
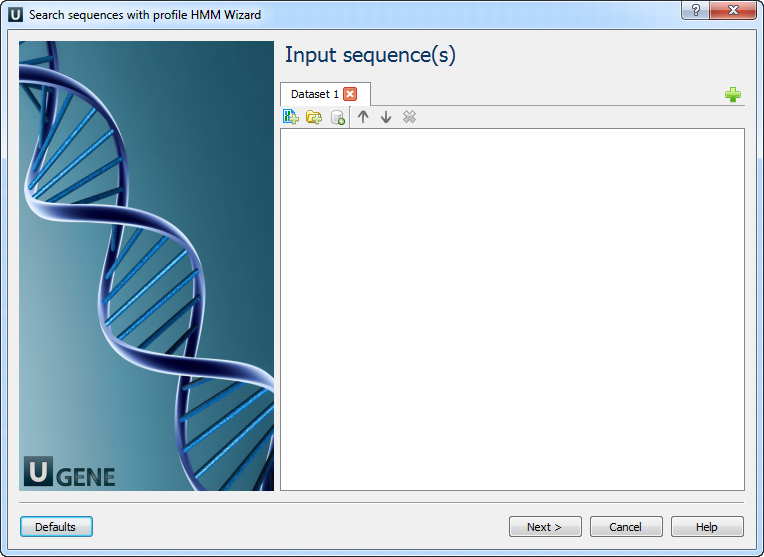
The wizard comprises two pages:
Input sequence(s): On this page, you must input sequence(s).
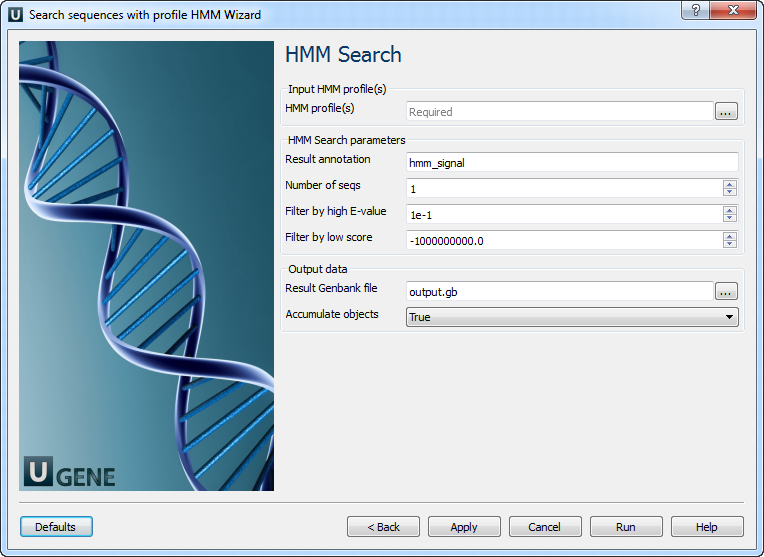
HMM search: On this page, you can modify HMM search parameters.
The following parameters are available:
HMM profile(s): A semicolon-separated list of paths to the input files.
Result annotation: A name for the result annotations.
Number of seqs: Calculate the E-value scores as if we had scanned a sequence database of sequences.
Filter by high E-value: E-value filtering can be used to exclude low-probability hits from the results.
Filter by low score: Score-based filtering is an alternative to E-value filtering to exclude low-probability hits from the results.
Result Genbank file: Location of the output data file. If this attribute is set, the “Location” slot in the port will not be used.
Accumulate objects: Accumulate all incoming data in one file or create separate files for each input. In the latter case, an incremental numerical suffix is added to the file name.