De novo Assemble Illumina PE Reads
The workflow sample, described below, takes FASTQ files with paired-end Illumina reads as input and processes them as follows:
- Improve reads quality with Trimmomatic
- Provide FastQC quality reports
- De novo assemble reads with SPAdes
How to Use This Sample
If you haven’t used the workflow samples in UGENE before, look at the “How to Use Sample Workflows” section of the documentation.
Workflow Sample Location
The workflow sample “De novo Assemble Illumina PE Reads” can be found in the “NGS” section of the Workflow Designer samples.
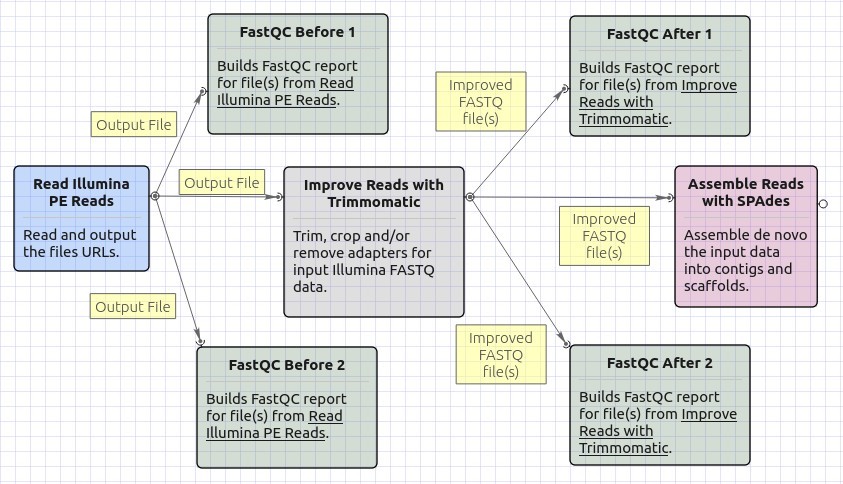
Workflow Image
The opened workflow looks as follows:
Workflow Wizard
The wizard has 4 pages.
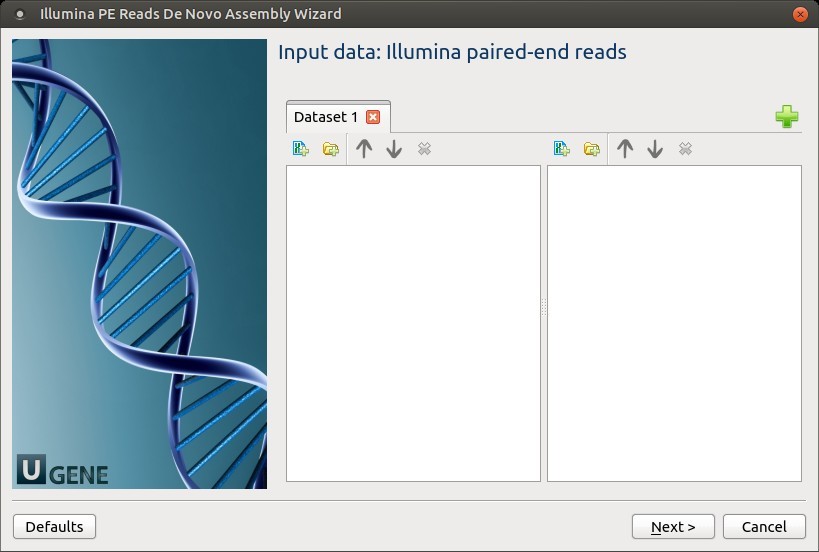
1. Input Data: Illumina Paired-End Reads
On this page, files with Illumina paired-end reads must be set.
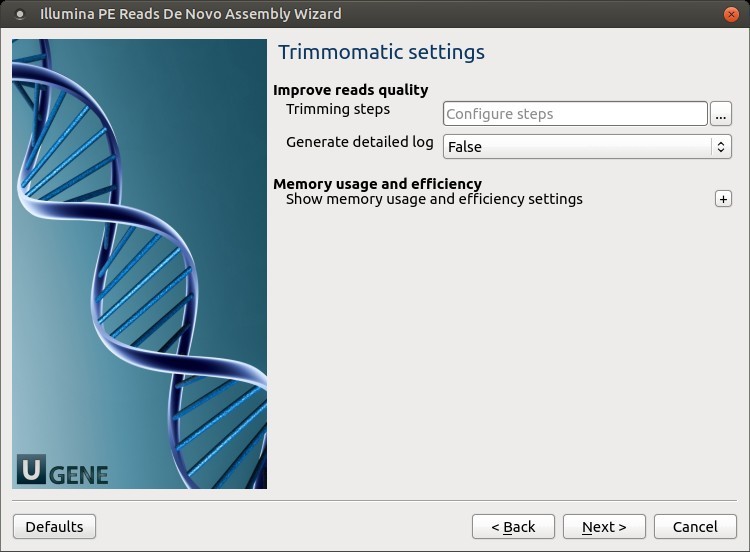
2. Trimmomatic Settings
The Trimmomatic parameters can be changed here:
To configure trimming steps, use the following button:
The following dialog will appear:
Click the Add new step button and select a step. The following options are available:
- ILLUMINACLIP
- SLIDINGWINDOW
- LEADING
- TRAILING
- CROP
- HEADCROP
- MINLEN
- AVGQUAL
- TOPHRED33
- TOPHRED64
Each step has its own parameters (see the original document for detailed explanations).
To remove a step, use the Remove selected step button. The pink highlighting means the required parameter has not been set.
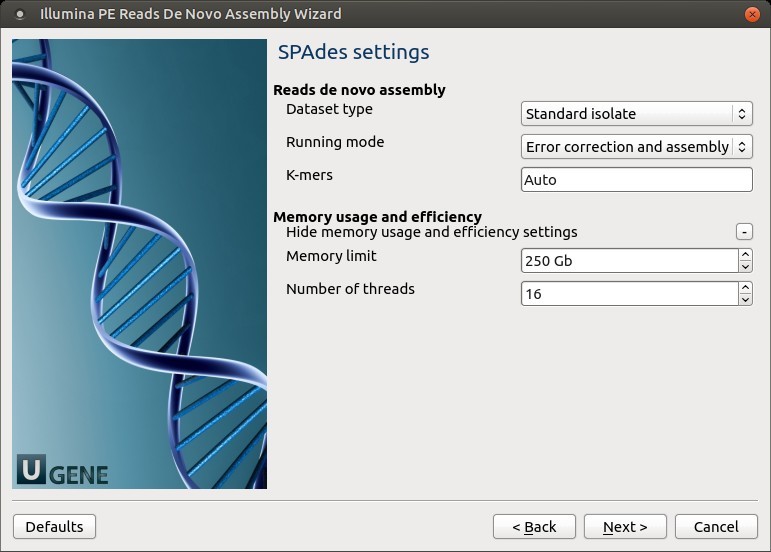
3. SPAdes Settings
Default SPAdes parameters can be changed here.
Parameters Table
| Parameter | Description |
|---|---|
| Dataset type | Select the input dataset type: - Standard isolate (default) - Multiple displacement amplification ( --sc) |
| Running mode | By default, SPAdes performs both read error correction and assembly. You can select: - --only-assembler (assembly only)- --only-error-correction (error correction only) |
| Error correction | SPAdes uses: - BayesHammer for Illumina - IonHammer for IonTorrent ⚠️ Do not use error correction with reads lacking quality info (e.g., FASTA) |
| K-mers | Specify -k k-mer sizes. If not set, SPAdes selects them automatically based on read length |
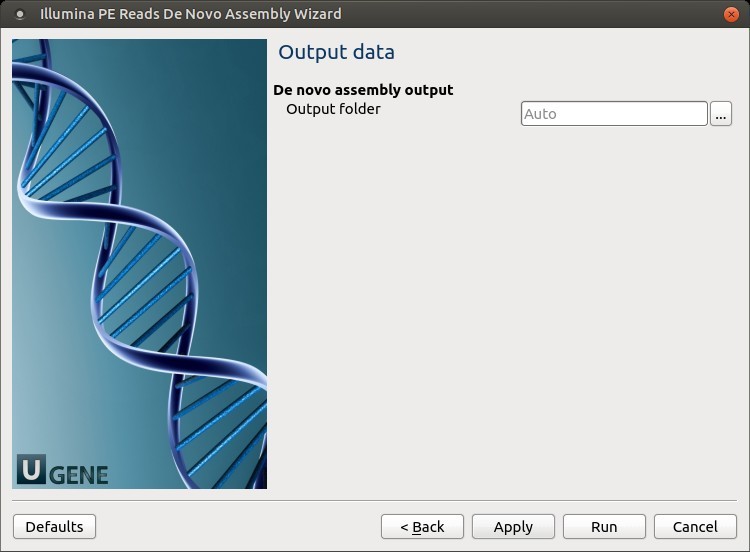
4. Output Files Page
On this page, you can select an output directory.