Parallel NGS Reads Classification
Attention: Metagenomics was available before v40.
The workflow sample described below takes FASTQ files with metagenomic NGS reads as input and processes them as follows:
- Improve reads quality with Trimmomatic
- Provide FastQC reads quality reports
- Classification:
- Classify the pre-processed reads with Kraken
- Classify the assembled contigs with CLARK
- Classify these reads with DIAMOND (in the case of SE reads)
- Classify these reads with MetaPhlAn2
- Ensemble the classification output from Kraken, CLARK, and DIAMOND into a CSV file.
- Improve classification from these tools with WEVOTE.
- Provide general classification reports
How to Use This Sample
If you haven’t used the workflow samples in UGENE before, refer to the “How to Use Sample Workflows” section of the documentation.
Workflow Sample Location
The workflow sample “Parallel NGS Reads Classification” can be found in the “NGS” section of the Workflow Designer samples.
Workflow Image
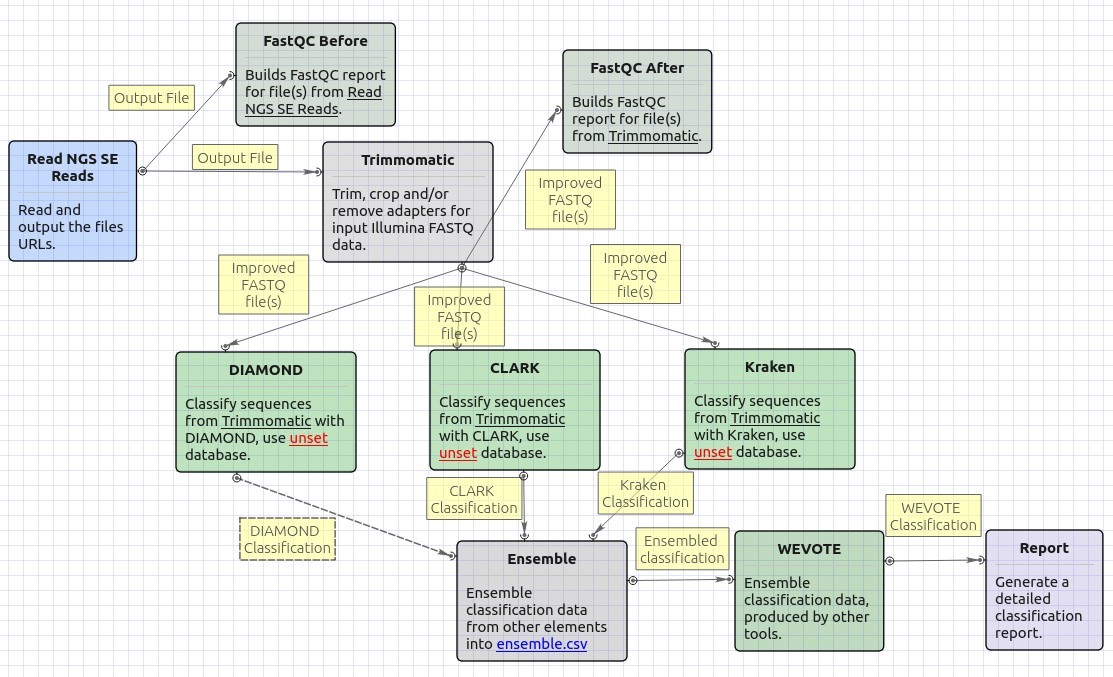
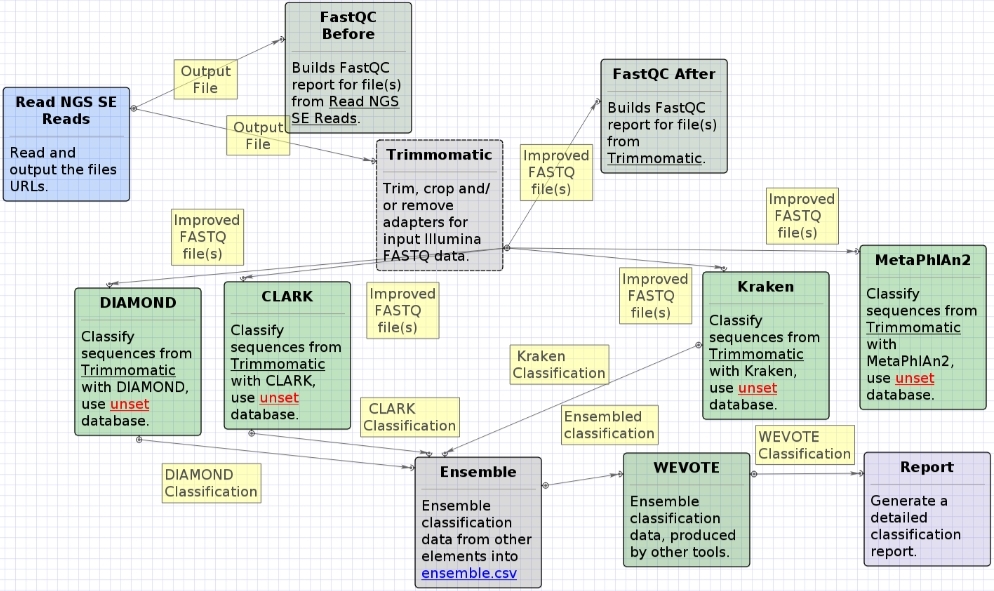
The opened workflow for single-end reads looks as follows:
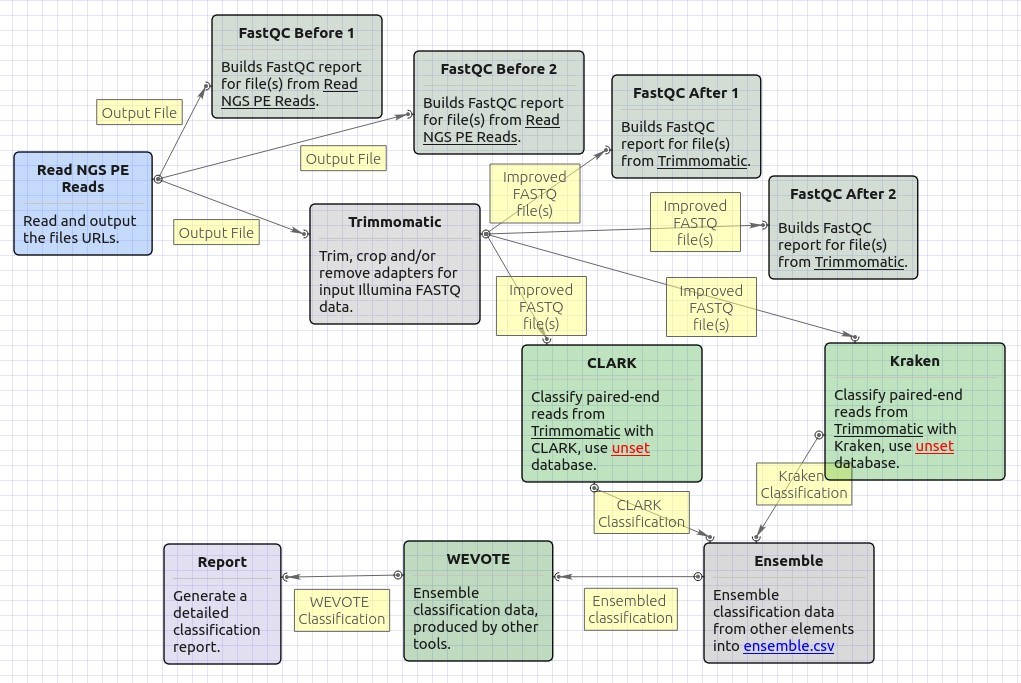
The opened workflow for paired-end reads looks as follows:
Workflow Wizard
The wizard has six pages.
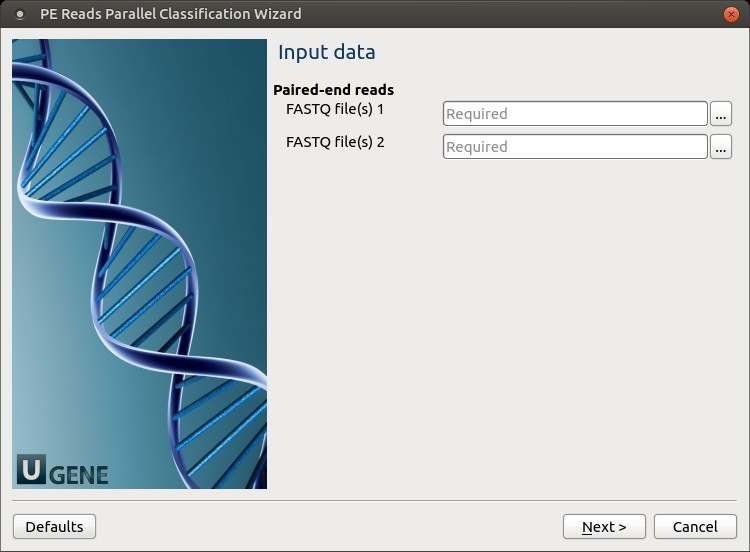
Input data: On this page, input files must be set.
Trimmomatic settings: The Trimmomatic parameters can be changed here.
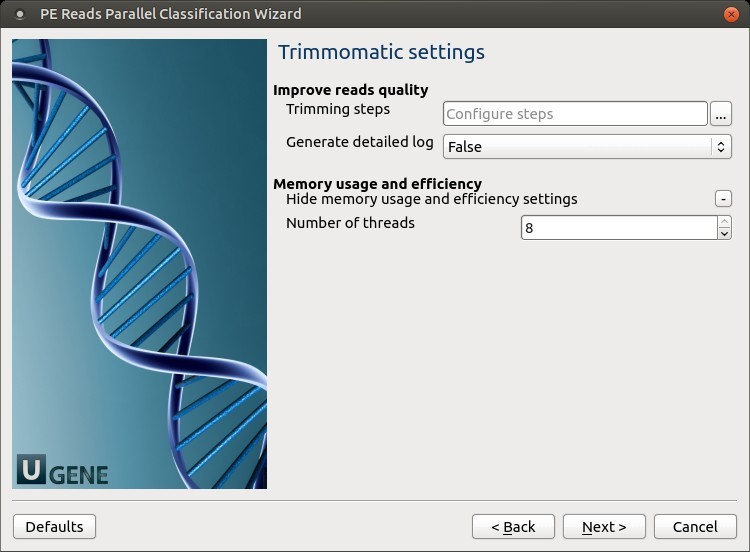
To configure trimming steps, use the following button:
The following dialog will appear:
Click the Add new step button and select a step. The following options are available:
- ILLUMINACLIP: Cut adapter and other Illumina-specific sequences from the read.
- SLIDINGWINDOW: Perform a sliding window trimming, cutting once the average quality within the window falls below a threshold.
- LEADING: Cut bases off the start of a read if below a threshold quality.
- TRAILING: Cut bases off the end of a read if below a threshold quality.
- CROP: Cut the read to a specified length.
- HEADCROP: Cut the specified number of bases from the start of the read.
- MINLEN: Drop the read if it is below a specified length.
- AVGQUAL: Drop the read if the average quality is below the specified level.
- TOPHRED33: Convert quality scores to Phred-33.
- TOPHRED64: Convert quality scores to Phred-64.
Each step has its own parameters:
AVGQUAL
This step drops a read if the average quality is below the specified level.
Input the following values:
- Quality threshold: the minimum average quality required to keep a read.
CROP
This step removes bases from the end of the read, ensuring it has at most the specified length after this step has been performed.
Input the following values:
- Length: the number of bases to keep, from the start of the read.
HEADCROP
This step removes the specified number of bases from the beginning of the read.
Input the following values:
- Length: the number of bases to remove from the start of the read.
ILLUMINACLIP
This step finds and removes Illumina adapters.
Trimmomatic first compares short sections of an adapter and a read. If they match sufficiently, the entire alignment between the read and adapter is scored. For paired-end reads, the “palindrome” approach is also used to improve the result. See the Trimmomatic manual for details.
Input the following values:
- Adapter sequences: a FASTA file with the adapter sequences. Files for TruSeq2 (GAII machines), TruSeq3 (HiSeq and MiSeq machines), and Nextera kits for SE and PE reads are available by default. The naming of the sequences within the specified file determines how they are used.
- Seed mismatches: the maximum mismatch count in short sections that still allows a full match to be performed.
- Simple clip threshold: a threshold for simple alignment mode. Values between 7 and 15 are recommended. A perfect match of a 12-base sequence scores just over 7, while 25 bases are needed to score 15.
- Palindrome clip threshold: a threshold for palindrome alignment mode. For palindromic matches, a longer alignment is possible. Therefore the threshold can be in the range of 30. Even though this threshold is very high (requiring a match of almost 50 bases), Trimmomatic can identify very short adapter fragments.
There are also two optional parameters for palindrome mode: Min adapter length and Keep both reads. Use the following dialog to set them. To access the dialog, press the Optional button.
LEADING
This step removes low-quality bases from the beginning of the read. As long as a base has a value below this threshold, the base is removed and the next base is examined.
Input the following values:
- Quality threshold: the minimum quality required to keep a base.
MAXINFO
This step performs an adaptive quality trim, balancing the benefits of retaining longer reads against the costs of retaining bases with errors. See the Trimmomatic manual for details.
Input the following values:
- Target length: the read length likely to allow locating the read within the target sequence. Extremely short reads, which can fit into many different locations, provide little value. Typically, the length would be around 40 bases. However, the value also depends on the size and complexity of the target sequence.
- Strictness: the balance between preserving as much read length as possible and removing incorrect bases. A low value of this parameter (0.8) favors read correctness.
MINLEN
This step removes reads falling below the specified minimum length. It should normally be placed after all other processing steps. Reads removed by this step will be counted and included in the “dropped reads” count.
Input the following values:
- Length: the minimum length of reads to be kept.
SLIDINGWINDOW
This step performs a sliding window trimming, cutting once the average quality within the window falls below a threshold. By considering multiple bases, a single poor-quality base will not cause the removal of high-quality data later in the read.
Input the following values:
- Window size: the number of bases to average across.
- Quality threshold: the average quality required.
TOPHRED33
This step re-encodes the quality part of the FASTQ file to base 33.
TOPHRED64
This step re-encodes the quality part of the FASTQ file to base 64.
TRAILING
This step removes low-quality bases from the end of the read. As long as a base has a value below this threshold, the base is removed and the next base (i.e., the preceding one) is examined. This approach can remove the special Illumina “low-quality segment” regions (marked with a quality score of 2), but SLIDINGWINDOW or MAXINFO are recommended instead.
Input the following values:
- Quality threshold: the minimum quality required to keep a base.
To remove a step, use the Remove selected step button. The pink highlighting means a required parameter has not been set.
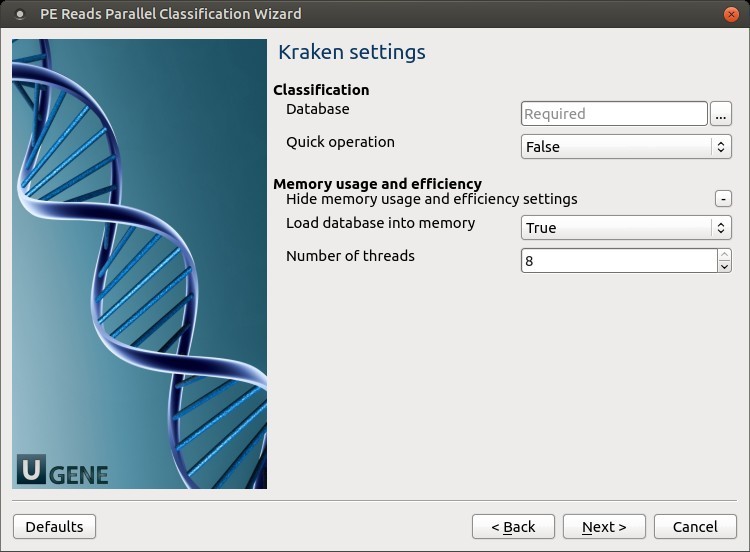
Kraken settings: Default Kraken parameters can be changed here.
The following parameters are available:
- Database: A path to the folder with the Kraken database files.
- Quick operation: Stop classification of an input read after a certain number of hits. This number can be specified in the “Minimum number of hits” parameter.
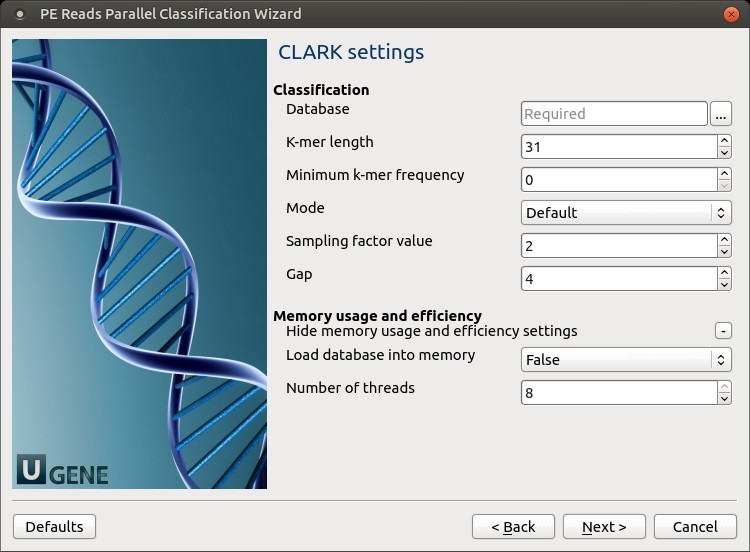
CLARK settings: Default CLARK parameters can be changed here.
The following parameters are available:
- Database: A folder that should be used to store the database files.
- K-mer length: This value is critical for classification accuracy and speed.
For high sensitivity, it is recommended to set this value to 20 or 21 (along with the “Full” mode). However, if precision and speed are the main concerns, use any value between 26 and 32. Note that the higher the value, the higher the RAM usage. As a good trade-off between speed, precision, and RAM usage, set this value to 31 (along with the “Default” or “Express” mode).
Minimum k-mer frequency: Minimum k-mer frequency/occurrence for the discriminative k-mers (-t). For example, for 1 (or, 2), the program will discard any discriminative k-mer appearing only once (or, less than twice).
Mode: Set the mode of execution (-m):
- “Full” to get detailed results, confidence scores, and other statistics.
- “Default” to get a results summary and perform the best trade-off between classification speed, accuracy, and RAM usage.
- “Express” to get a results summary with the highest speed possible.
Sampling factor value
Gap: “Gap” or number of non-overlapping k-mers to pass when creating the database (-п). Increase the value if it’s required to reduce RAM usage. Note that this will degrade sensitivity.
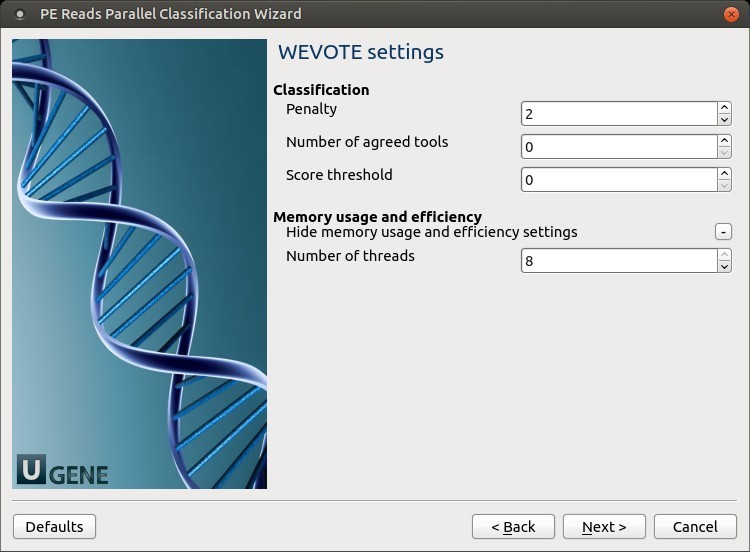
WEVOTE settings: Default WEVOTE parameters can be changed here.
The following parameters are available:
- Penalty: Score penalty for disagreements (-k)
- Number of agreed tools: Specify the minimum number of tools agreed on for WEVOTE decision (-a).
- Score threshold: Score threshold (-s)
Output Files Page: On this page, you can select an output directory.