Raw RNA-Seq Data Processing
Use this workflow sample to process raw RNA-seq next-generation sequencing (NGS) data from the Illumina platform. The processing includes:
- Filtration:
- Filtering of the NGS short reads by the CASAVA 1.8 header;
- Trimming of the short reads by quality;
- [Optionally] Mapping:
- Mapping of the short reads to the specified reference sequence (the TopHat tool is used in the sample);
The resulting output of the workflow contains the filtered and merged FASTQ files. In case the TopHat mapping has been done, the result also contains the TopHat output files: the accepted hits BAM file and tracks of junctions, insertions, and deletions in BED format. Other intermediate data files are also output by the workflow.
How to Use This Sample
If you haven’t used the workflow samples in UGENE before, look at the “How to Use Sample Workflows” section of the documentation.
What’s Next?
The Tuxedo workflow can be used to analyze the filtered RNA-seq data. In this case, the mapping step of this workflow can be skipped, as it is also present in the Tuxedo pipeline.
Workflow Sample Location
The workflow sample “Raw DNA-Seq processing” can be found in the “NGS” section of the Workflow Designer samples.
Workflow Image
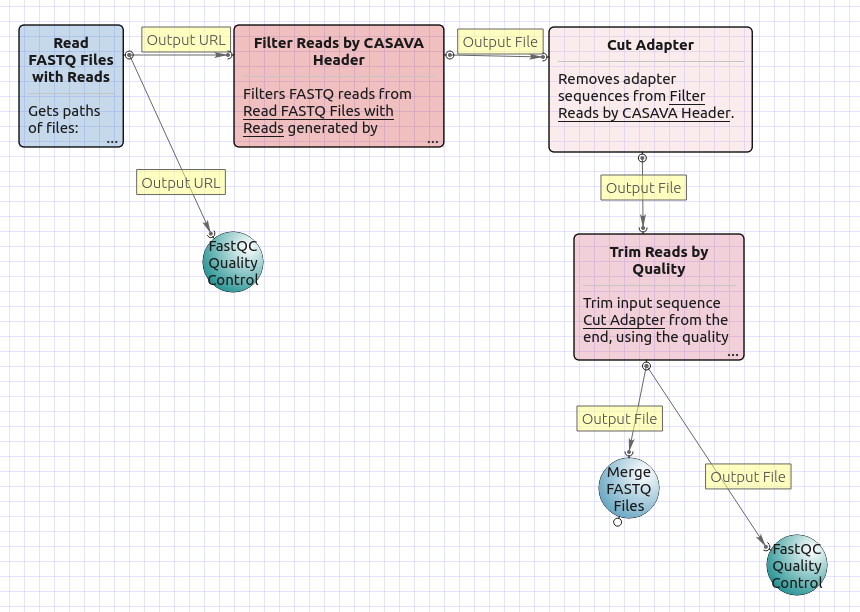
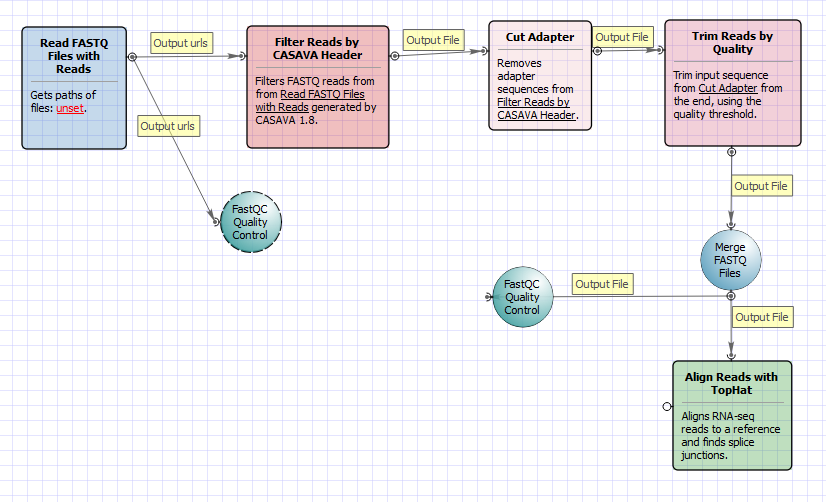
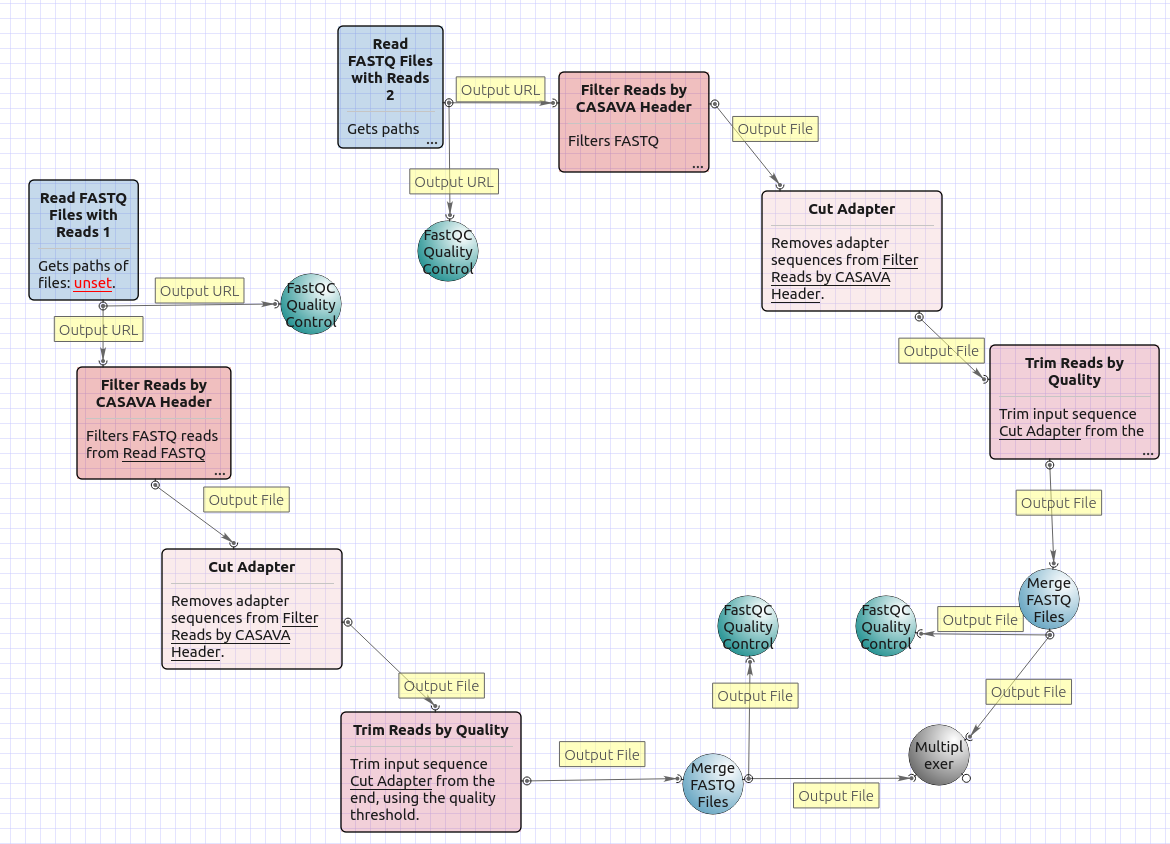
There are four versions of the workflow available. The workflow with mapping for single-end reads looks as follows:
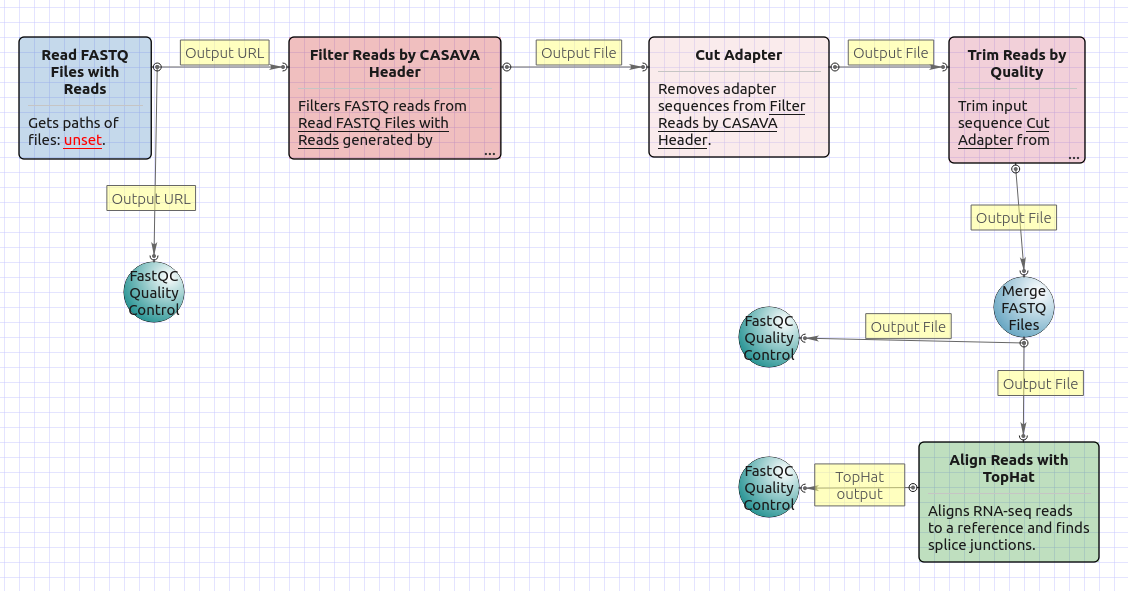
The workflow with mapping for paired-end reads appearance is as follows:
The workflow without mapping for single-end reads appearance is as follows:
The workflow without mapping for paired-end reads appearance is as follows:
Workflow Wizard
The workflows have similar wizards. The wizard for paired-end reads with mapping has four pages.
Input data: On this page, you must input FASTQ file(s).
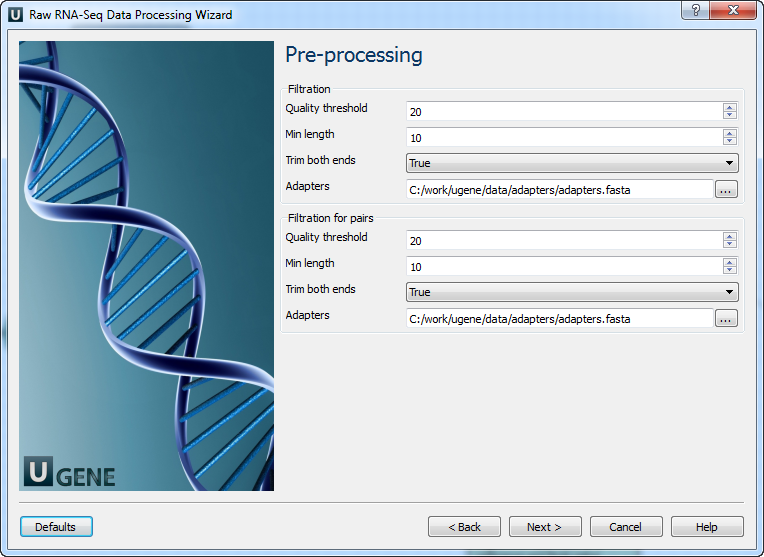
Pre-processing: On this page, you can modify filtration parameters.
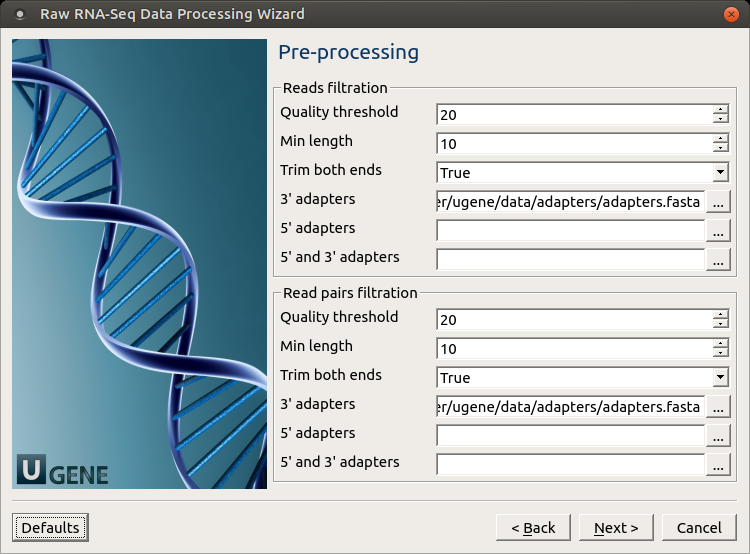
The following parameters are available for reads and reads pairs filtration:
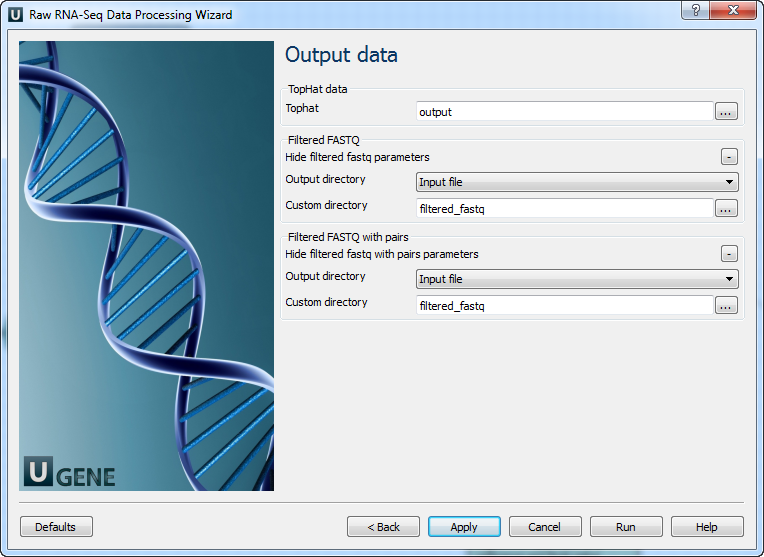
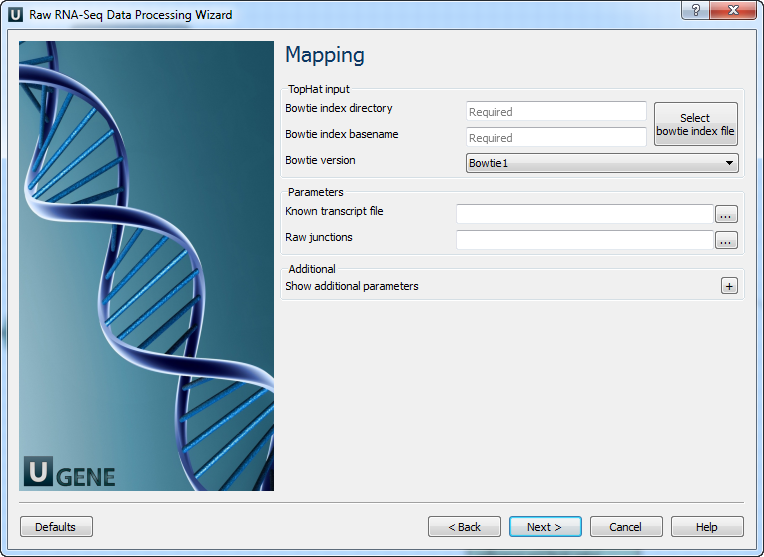
Parameter Description Base quality Quality threshold for trimming. Reads length Too short reads are discarded by the filter. Trim both ends Trim both ends of a read or not. Usually set to True for Sanger sequencing and False for NGS 3’ adapters A FASTA file with sequences of adapters that were ligated to the 3’ end. The adapter and any sequence following it are trimmed. If the adapter sequence ends with the ‘$’ character, it is anchored to the end of the read and only found as a suffix. 5’ adapters A FASTA file with sequences of adapters that were ligated to the 5’ end. If the adapter sequence starts with the ‘^’ character, it is ‘anchored’. Anchored adapters must appear entirely at the 5’ end. Non-anchored may appear partially. 5’ and 3’ adapters A FASTA file with sequences of adapters that were ligated to either the 5’ end or 3’ end. Mapping: On this page, you must input reference and optionally modify advanced parameters.
The following parameters are available:
Parameter Description Bowtie index directory The directory with the Bowtie index for the reference sequence. Bowtie index basename The basename of the Bowtie index for the reference sequence. Bowtie version Specifies which Bowtie version should be used. Known transcript file A set of gene model annotations and/or known transcripts. Raw junctions The list of raw junctions. Mate inner distance Expected (mean) inner distance between mate pairs. Mate standard deviation Standard deviation for the distribution of inner distances between mate pairs. Library type Specifies RNA-seq protocol. No novel junctions Only search for reads across junctions indicated in the supplied GFF or junctions file. Ignored if ‘Raw junctions’ or ‘Known transcript file’ is not set. Max multihints Allows up to this many alignments to the reference for a given read, suppressing all alignments for reads with more than this many. Segment length Each read is cut up into segments, at least this long, mapped independently. Fusion search Turn on fusion mapping. Transcriptome max hits Align reads to the transcriptome and report only those mappings as genomic mappings. Prefilter multihints Align reads to the genome first to exclude multi-mapped reads appearing to align to transcript sequences due to repetitive or low complexity. Min anchor length Junctions spanned by reads with at least this many bases on each side are reported. Splice mismatches Maximum number of mismatches allowed in the anchor region of a spliced alignment. Read mismatches Final read alignments having more than these mismatches are discarded. Segment mismatches Read segments mapped independently allowing up to these mismatches in each segment. Solexa 1.3 quals Use this option for FASTQ files from Illumina GA pipeline version 1.3 or later, where quality scores are Phred-scaled base-64. Bowtie -n mode Using Bowtie -n instead of -v for initial read mapping. Read segments use -v option. Bowtie tool path Path to the Bowtie external tool. SAMtools tool path Path to the SAMtools tool, available in UGENE External Tool Package. TopHat tool path Path to the TopHat external tool in UGENE. Temporary directory Directory for temporary files. Output data: On this page, you must input output parameters.