RNA-seq Analysis with Tuxedo Tools
The RNA-seq pipeline “Tuxedo” consists of the TopHat spliced read mapper, which internally uses Bowtie or Bowtie 2 short read aligners, and several Cufflinks tools that allow for the assembly of transcripts, estimation of their abundances, and testing for differential expression and regulation in RNA-seq samples.
Environment Requirements
The pipeline is currently available on Linux and macOS systems only.
How to Use This Sample
If you haven’t used the workflow samples in UGENE before, look at the “How to Use Sample Workflows” section of the documentation.
Workflow Sample Location
The workflow sample “RNA-seq Analysis with Tuxedo Tools” can be found in the “NGS” section of the Workflow Designer samples.
Workflow Image
There are two types of short read workflows: single-end and paired-end reads. For both of them, there are three analysis types:
- Full Tuxedo Pipeline - use this pipeline to analyze multiple samples with TopHat, Cufflinks, Cuffmerge, and Cuffdiff tools.
- Single-sample Tuxedo Pipeline - use this pipeline to analyze a single sample with TopHat and Cufflinks tools.
- No-new-transcripts Tuxedo Pipeline - use this pipeline to analyze multiple samples with TopHat and Cuffdiff tools only, i.e., without producing new transcripts.
For the Full Tuxedo Pipeline analysis type and single-end reads type, the following workflow appears:
For the Full Tuxedo Pipeline analysis type and paired-end reads type, the following workflow appears:
For the Single-sample Tuxedo Pipeline analysis type and single-end reads type, the following workflow appears:
For the Single-sample Tuxedo Pipeline analysis type and paired-end reads type, the following workflow appears:
For the No-new-transcripts Tuxedo Pipeline analysis type and single-end reads type, the following workflow appears:
For the No-new-transcripts Tuxedo Pipeline analysis type and paired-end reads type, the following workflow appears:
Workflow Wizard
All of these workflows have similar wizards. For the Full Tuxedo Pipeline analysis type and the paired-end reads type, the wizard has 7 pages.
Input Data: Here, you need to input RNA-seq short reads in FASTA or FASTQ formats. Multiple datasets with different reads can be added.
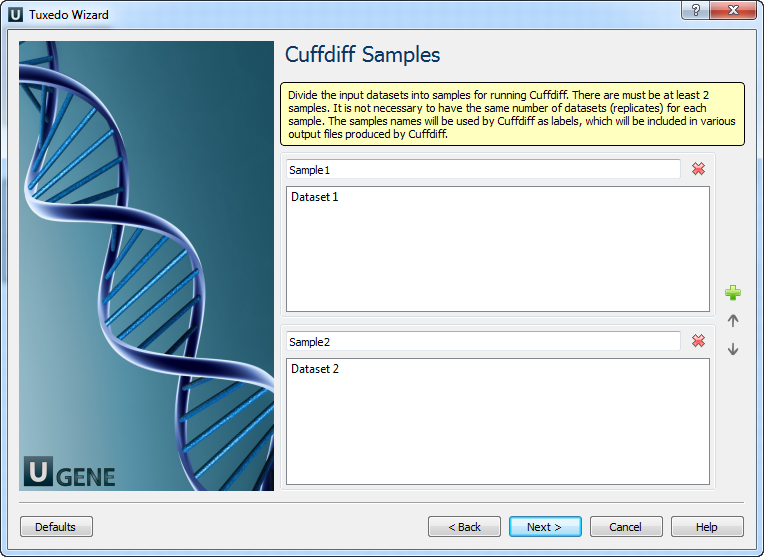
Cuffdiff Samples: Here, you need to divide the input datasets into samples for running Cuffdiff. There must be at least 2 samples. It is not necessary to have the same number of datasets (replicates) for each sample. The sample names will be used by Cuffdiff as labels, which will be included in various output files produced by Cuffdiff.
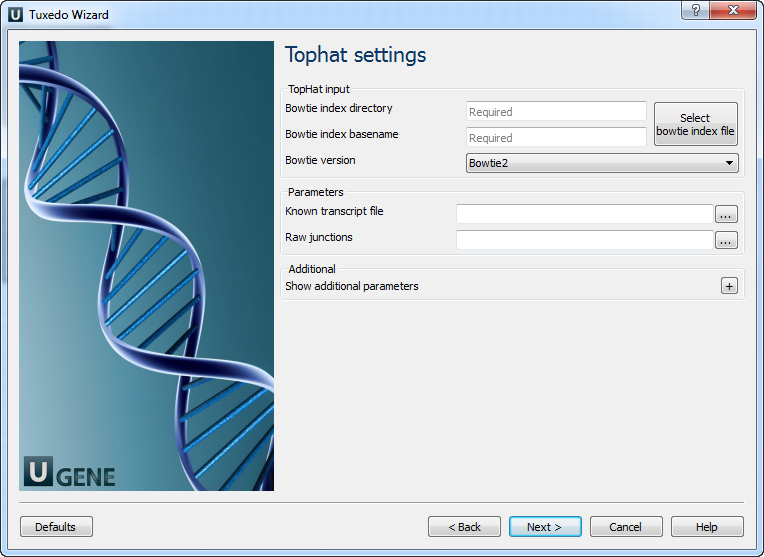
TopHat Settings: Here, you can configure TopHat settings. To show additional parameters, click on the + button.
The following parameters are available:
Parameter Description Bowtie index directory The directory with the Bowtie index for the reference sequence. Bowtie index basename The basename of the Bowtie index for the reference sequence. Bowtie version Specifies which Bowtie version should be used. Known transcript file A set of gene model annotations and/or known transcripts. Raw junctions The list of raw junctions. Mate inner distance Expected (mean) inner distance between mate pairs. Mate standard deviation Standard deviation for the distribution on inner distances between mate pairs. Library type Specifies RNA-seq protocol. No novel junctions Only look for reads across junctions indicated in the supplied GFF or junctions file. (Ignored if Raw junctions or Known transcript file is not set.) Max multihits Allows up to this many alignments to the reference for a given read, and suppresses all alignments for reads with more than this many alignments. Segment length Each read is cut up into segments, each at least this long. These segments are mapped independently. Fusion search Turn on fusion mapping. Transcriptome max hits Only align the reads to the transcriptome and report only those mappings as genomic mappings. Prefilter multihits Directs TopHat to first align the reads to the whole genome and exclude multi-mapped reads based on the Max multihits option value. Min anchor length The anchor length. TopHat will report junctions spanned by reads with at least this many bases on each side of the junction. Splice mismatches The maximum number of mismatches that may appear in the anchor region of a spliced alignment. Read mismatches Final read alignments having more than this many mismatches are discarded. Segment mismatches Read segments are mapped independently, allowing up to this many mismatches in each segment alignment. Solexa 1.3 quals Use for FASTQ files from pipeline 1.3 or later, where quality scores are encoded in Phred-scaled base-64. Bowtie -n mode Use -n in Bowtie for initial read mapping. Read segments are always mapped using -v option. Bowtie tool path The path to the Bowtie external tool. SAMtools tool path The path to the SAMtools tool. (Note that the tool is available in the UGENE External Tool Package.) TopHat tool path The path to the TopHat external tool in UGENE. Temporary directory The directory for temporary files. Cufflinks Settings: The following page allows you to configure Cufflinks settings:
The following parameters are available:
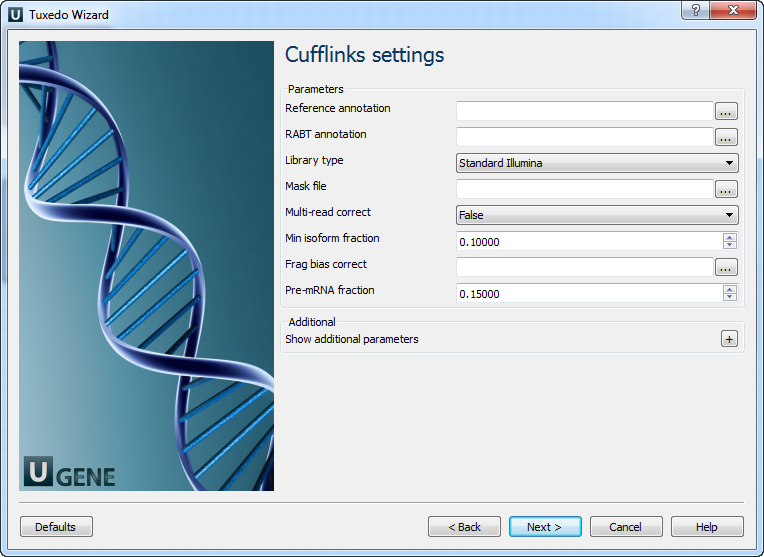
Parameter Description Reference annotation Tells Cufflinks to use the supplied reference annotation to estimate isoform expression. RABT annotation Use the supplied reference annotation to guide Reference Annotation Based Transcript (RABT) assembly. Library type Specifies RNA-seq protocol. Mask file Ignore all reads that could have come from transcripts in this file. Multi-read correct More accurately weight reads mapping to multiple locations in the genome. Min isoform fraction Filters out transcripts with very low abundance. Frag bias correct Providing Cufflinks with a multifasta file instructs it to run the bias detection and correction algorithm. Pre-mRNA fraction Filters out alignments in intronic intervals implied by spliced alignments. Cufflinks tool path The path to the Cufflinks external tool in UGENE. Temporary directory The directory for temporary files. Cuffmerge Settings: On this page, you can modify Cuffmerge parameters:
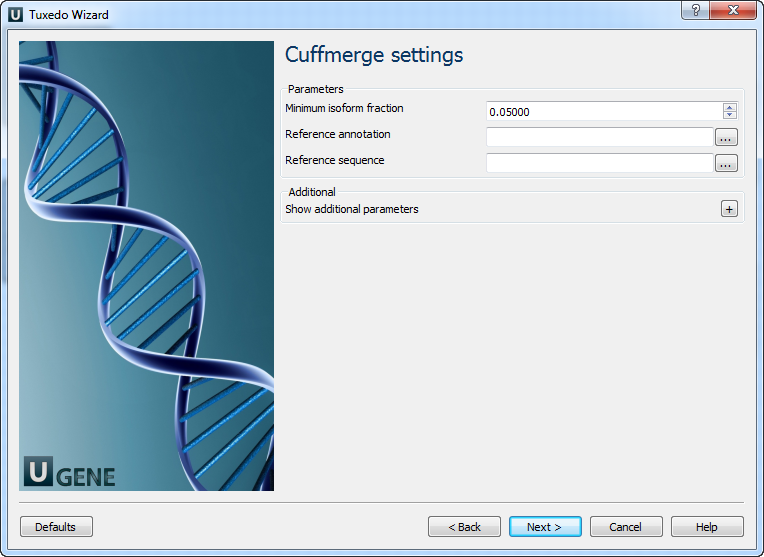
The following parameters are available:
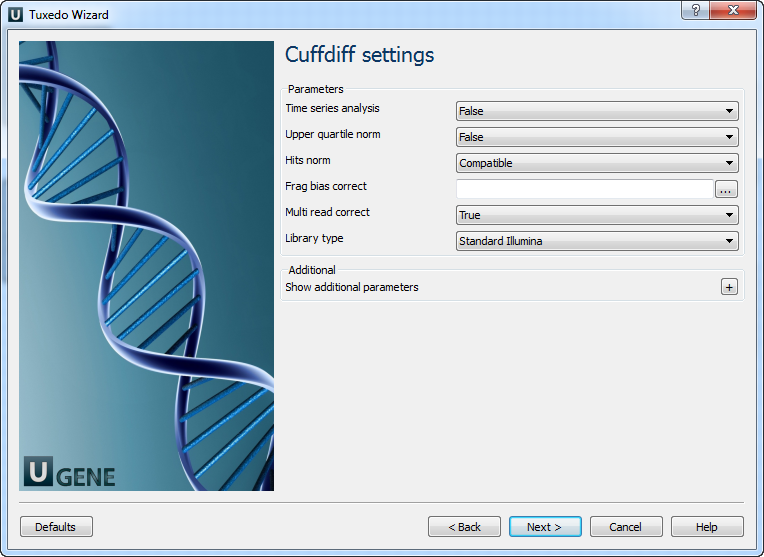
Parameter Description Minimum isoform fraction Discard isoforms with abundance below this. Reference annotation Merge the input assemblies together with this reference annotation. Reference sequence The genomic DNA sequences for the reference. Used to classify transfrags and exclude artifacts. Cuffcompare tool path The path to the Cuffcompare external tool in UGENE. Cuffmerge tool path The path to the Cuffmerge external tool in UGENE. Temporary directory The directory for temporary files. Cuffdiff Settings: On the following page, you may configure Cuffdiff settings:
The following parameters are available:
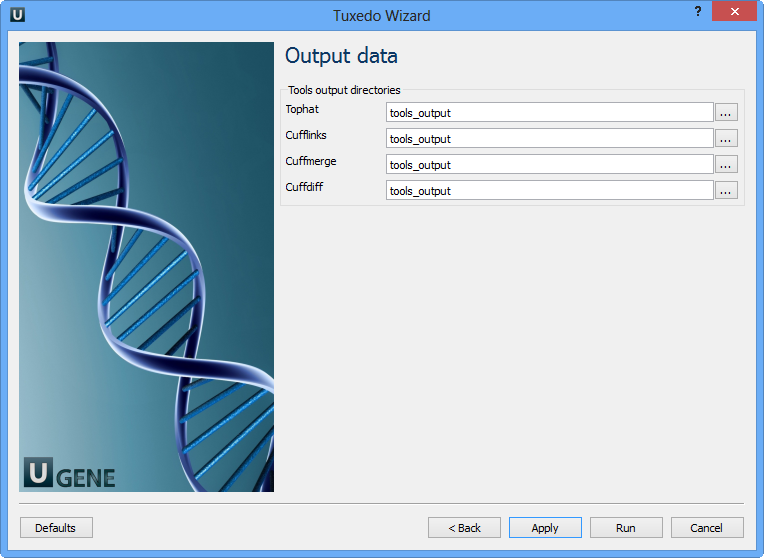
Parameter Description Time series analysis Instructs Cuffdiff to analyze the samples as a time series. Upper quartile norm Normalizes by the upper quartile of the number of fragments mapping to individual loci. Hits norm Instructs how to count all fragments. Choosing Compatible is generally recommended to reduce bias. Frag bias correct Provides sequences to Cuffdiff for bias detection and correction. Multi-read correct More accurately weight reads mapping to multiple locations in the genome. Library type Specifies RNA-seq protocol. Mask file Ignore all reads that could have originated from transcripts in this file. Min alignment count The minimum number of alignments in a locus necessary for significance testing. FDR Allowed false discovery rate used in testing. Max MLE iterations Sets the maximum number of iterations during maximum likelihood estimation of abundances. Emit count tables Include information about fragment counts and variances in the report. Cuffdiff tool path The path to the Cuffdiff external tool in UGENE. Temporary directory The directory for temporary files. Output Data: On this page, you can modify output parameters.
The work on this pipeline was supported by grant RUB1-31097-NO-12 from NIAID.