Gene-by-gene Approach for Characterization of Genomes
Suppose you have genomes and you want to characterize them. One of the ways to do that is to build a table detailing which genes are present in each genome and which are not.
Step-by-step Instructions
- Create a local BLAST database of your genome sequence/contigs. One database per genome.
- Create a file with sequences of the genes you want to explore. This file will serve as the input file for the workflow.
- Set up the location and name of the BLAST database you created for the first genome.
- Set up output files: report location and (optionally) a file with annotated (via BLAST) sequences. You can delete the Write Sequence element if output sequences are not needed.
- Run the workflow.
- Run the workflow again on the same input and output files, changing only the BLAST database for each genome you have.
As a result, you’ll get a report file with “Yes” and “No” values:
- “Yes” means the gene is found in the genome.
- “No” might mean the gene is missing, but it’s recommended to analyze all the “No” cases using the annotated files. Just open the file and find the sequence by gene name.
How to Use This Sample
If you haven’t used workflow samples in UGENE before, refer to How to Use Sample Workflows.
Workflow Sample Location
The workflow sample Gene-by-gene Approach for Characterization of Genomes is in the Scenarios section of the Workflow Designer samples.
Workflow Image
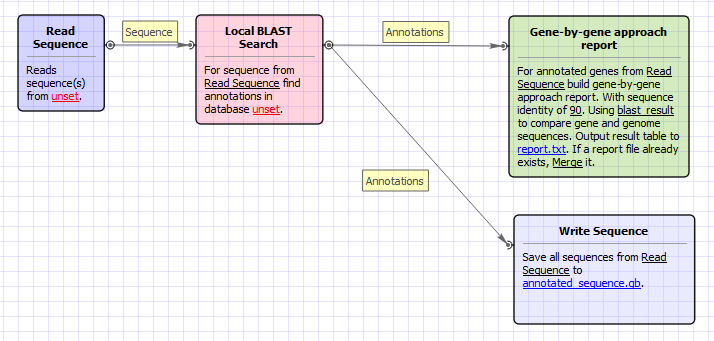
The workflow looks as follows:
Workflow Wizard
The wizard includes three pages:
1. Input Sequence(s)
On this page, you must input sequence(s):
2. BLAST Search Settings
On this page, you can configure BLAST search parameters:
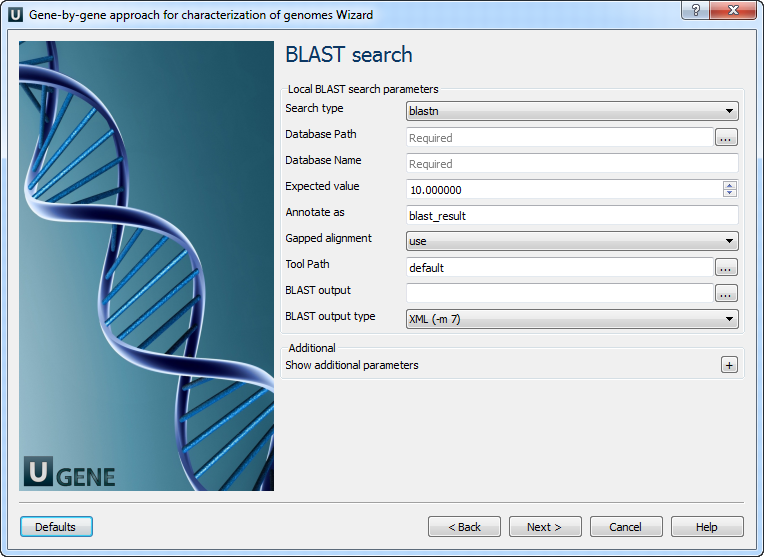
| Parameter | Description |
|---|---|
| Search type | Select the type of BLAST search. |
| Database Path | Path to the folder with BLAST database files. |
| Database Name | Base name for BLAST database files. |
| Expected value | Statistical threshold for reporting matches (E-value). |
| Annotate as | Name to use for result annotations. |
| Gapped alignment | Whether to perform gapped alignment. |
| Tool Path | Path to the external BLAST tool. |
| BLAST output | Path to the BLAST result file. |
| BLAST output type | Format of the BLAST result file. |
| Temporary directory | Location to store temporary files. |
| Gap costs | Cost for creating/extending a gap. |
| Match scores | Match reward and mismatch penalty. |
3. Output Data
On this page, you can set output options: