Search for Transcription Factor Binding Sites (TFBS) in Genomic Sequences
This workflow predicts binding sites for a number of transcription factors of interest using the SITECON algorithm. The current workflow sample is designed for the simultaneous recognition of binding sites for three different transcription factor types. You can expand it for the recognition of any desired number of transcription factor types. SITECON is a program package for the recognition of potential transcription factor binding sites based on the data about conservative conformational and physicochemical properties revealed from the analysis of binding site sets.
Citing SITECON: Please cite: Oshchepkov D.Y., Vityaev E.E., Grigorovich D.A., Ignatieva E.V., Khlebodarova T.M. SITECON: a tool for detecting conservative conformational and physicochemical properties in transcription factor binding site alignments and for site recognition. // Nucleic Acids Res. 2004 Jul 1;32(Web Server issue):W208-12.
How to Use This Sample
If you haven’t used the workflow samples in UGENE before, check the “How to Use Sample Workflows” section of the documentation.
Workflow Sample Location
The workflow sample “Search for Transcription Factor Binding Sites (TFBS) in Genomic Sequences” can be found in the “Transcriptomics” section of the Workflow Designer samples.
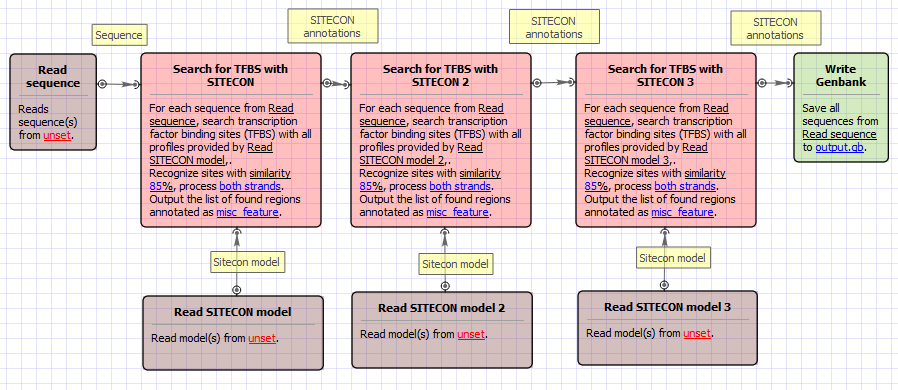
Workflow Image
The workflow looks as follows:
Workflow Wizard
The wizard has 5 pages.
Input sequence(s): On this page, you must input sequence(s).
Search for TFBS 1, 2, 3: On these pages, you can modify search for TFBS parameters.
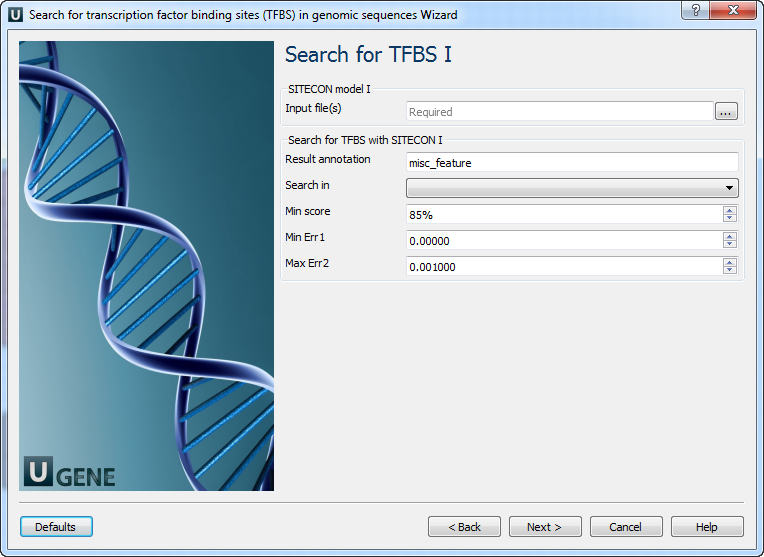
The following parameters are available:
Parameter Description Input file(s) Semicolon-separated list of paths to the input files. Result annotation Annotation name for marking found regions. Search in Which strands should be searched: direct, complement, or both. Min score Recognition quality percentage threshold. To switch off this filter, choose the lowest value. Min Err 1 Alternative setting for filtering results. Minimal value of Error type I. Note that all thresholds (by score, by Err1, and by Err2) are applied when filtering results. To switch off this filter, choose “0” value. Max Err 2 Alternative setting for filtering results. Max value of Error type II. Note that all thresholds (by score, by Err1, and by Err2) are applied when filtering results. To switch off this filter, choose “1” value. Output data: On this page, you can modify the output parameters.