Tutorial: NCBI BLAST Search Feature of UGENE Workflow Designer.
BLAST search against NCBI probably one of the most popular features in bioinformatics. There are many services, including NCBI itself for BLAST search. This tutorial presents the UGENE version of BLAST search. If you have multiple sequences to do a BLAST search probably you do not want to check them one by one. If you want to do a BLAST search for multiple sequences in a batch this tutorial might help you.
UGENE Concepts
There are a few UGENE concepts them you need to understand to do UGENE BLAST smoothly. If you are familiar with them, just skip this section.
1. UGENE BLAST results are represented as annotations on a sequence. So what you will get as a result of your BLAST search is an original sequence with similar BLAST regions that are saved a annotations. There is a way to see the original BLAST parameters and sequences and download the sequence using the qualifiers of an annotation.
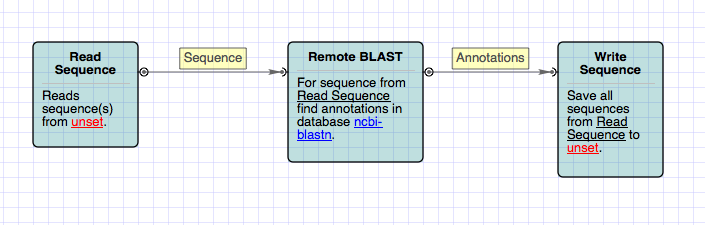
2. UGENE Workflow Designer allows you to do batch processing. You create a computational scheme that applies the same task to multiple datasets. If you create a scheme with a BLAST element it will apply the BLAST search to as many input sequences as you specify. The description of such a scheme is below.
Remote BLAST in Workflow Designer
UGENE Workflow Designer provides the ready-to-use sample for running NCBI BLAST search. Open Workflow Designer and find the “Remote BLASTing” sample. This sample has a wizard that makes adjusting of the workflow more easy and comfortable. We will describe the wizard windows further.
You can add several nucleotide sequences as an input of the workflow and each sequence will be processed separately.
Remote NCBI BLAST search has a set of parameters available in the second page of the wizard. For example, you can change the maximum number of results for all input sequences or switch off the “Megablast” option for making the search process more thorough. Note when you run the scheme, the remote BLAST element will make a call to NCBI service for each sequence. So you need an Internet connection for that workflow.
The result file names will be generated automatically.
For each input sequence UGENE creates a directory with two files: a file with the original sequence annotated with found BLAST results; and a file with the target BLAST alignments. The names of the directories are the same as the names of the files with the input sequences.
Click a file link to open the file with UGENE. You can see the resulting sequences with BLAST annotations now.

