3D Structures and Surface Algorithms
3D Structures (3D Structure Viewer). The 3D Structure Viewer is designed for the visualization of 3D structures of biological molecules. Using the 3D Structure Viewer, you can work with data from the Protein Data Bank (PDB), a repository for the 3D structural data of large biological molecules, such as proteins and nucleic acids, maintained by the Worldwide Protein Data Bank (wwPDB). You can also work with data from the NCBI Molecular Modeling Database (MMDB), also known as “Entrez Structure,” a database of experimentally determined structures obtained from the RCSB Protein Data Bank.
The 3D Structure Viewer opens automatically when you open a PDB or MMDB file. For example, open $ ugene/data/samples/PDB/1CF7.PDB. The 3D Structure Viewer adds a view to the upper part of the Sequence View:
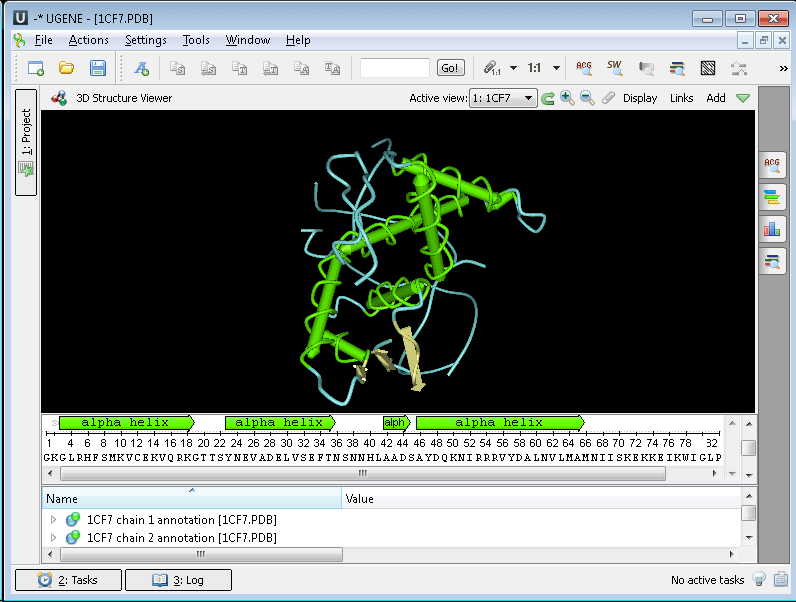
Using the 3D Structure Viewer, you can:
- Change 3D Structure Appearance.
- Move, zoom, and spin 3D structures.
- Highlight sequence regions.
- Select models to display.
- Export images of 3D structures.
- Work with several 3D structure views.
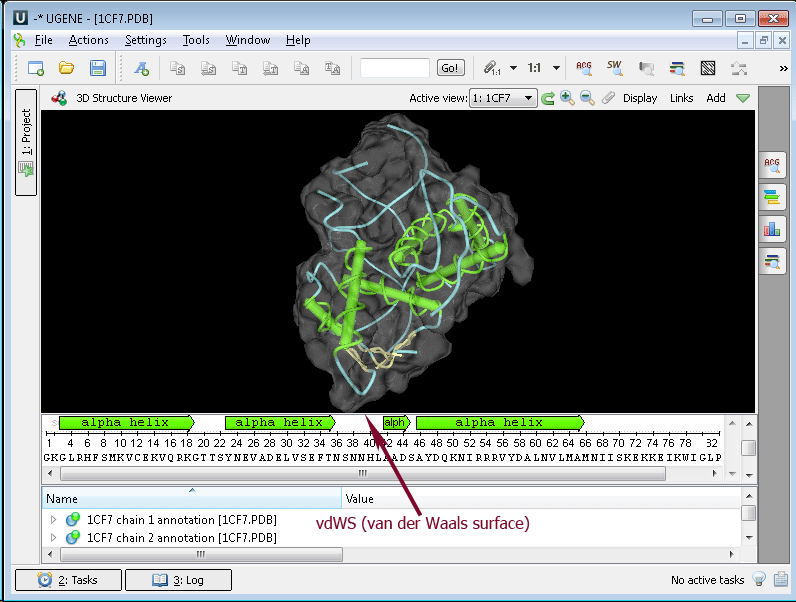
Example 5: Calculating Molecular Surface. To calculate the molecular surface of a molecule, select the Molecular Surface item in the 3D Structure Viewer context menu or in the Display menu on the toolbar and check one of the following items:
- SAS (solvent-accessible surface)
- SES (solvent-excluded surface)
- vdWS (van der Waals surface)
To remove an already calculated molecular surface, select the Off item. You can also select the Molecular Surface Render Style to modify the calculated molecular surface appearance:
- Convex Map
- Dots