View, Edit, and Annotate DNA, RNA, and Protein Sequences
DNA, RNA, or protein sequences (Sequence View). The Sequence View is one of the major object views in UGENE designed to visualize and edit DNA, RNA, or protein sequences along with their properties like annotations, chromatograms, 3D models, statistical data, etc. For each file, UGENE analyzes the file content and automatically opens the most appropriate view. To activate the Sequence View, open any file with at least one sequence. For example, you can use the $ugene/data/samples/Genbank/murine.gb file provided with UGENE. After opening the file in UGENE, the Sequence View window appears:
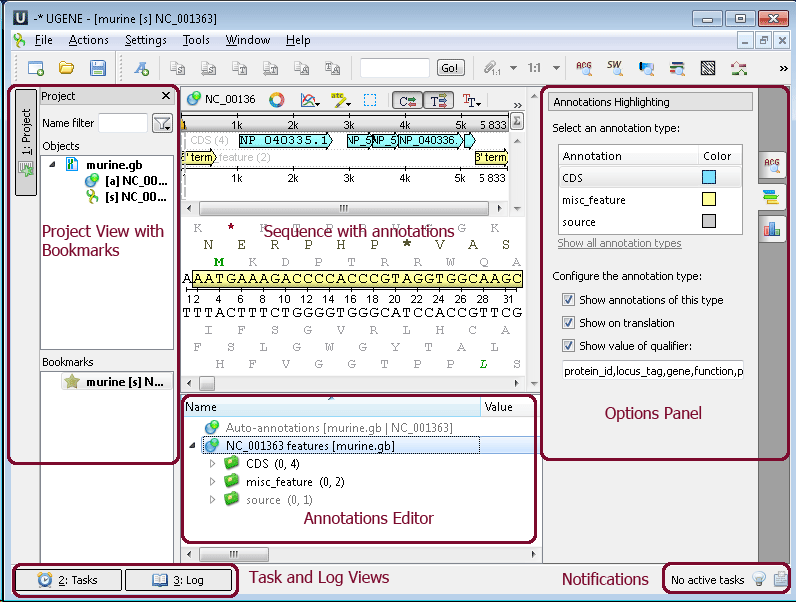
Sequence View - an object view designed to visualize DNA, RNA, or protein sequences along with their properties like annotations, chromatograms, 3D models, statistical data, etc.
Project View - a visual component used to manage active projects and bookmarks.
Annotations Editor - contains tools to manipulate annotations for a sequence.
Options Panel - a panel with different information tabs and tabs with settings for Sequence View.
Task View - shows active tasks, for example, algorithm computations.
Log View - shows the program log information.
Notifications - shows notifications for task reports.
After the view is opened, you can see a set of new buttons in the toolbar area. The actions provided by these buttons are available for all sequences opened in the view. These actions are also available from the context menu. Many instruments and algorithms are available:
- Find pattern
- Find ORFs
- Find annotated regions
- Build dotplot
- Find repeats
- Find tandems
- Find restriction sites
- Primer 3
- Find high DNA flexibility regions
- …
Example 1: Finding patterns in your sequence. Do the following steps:
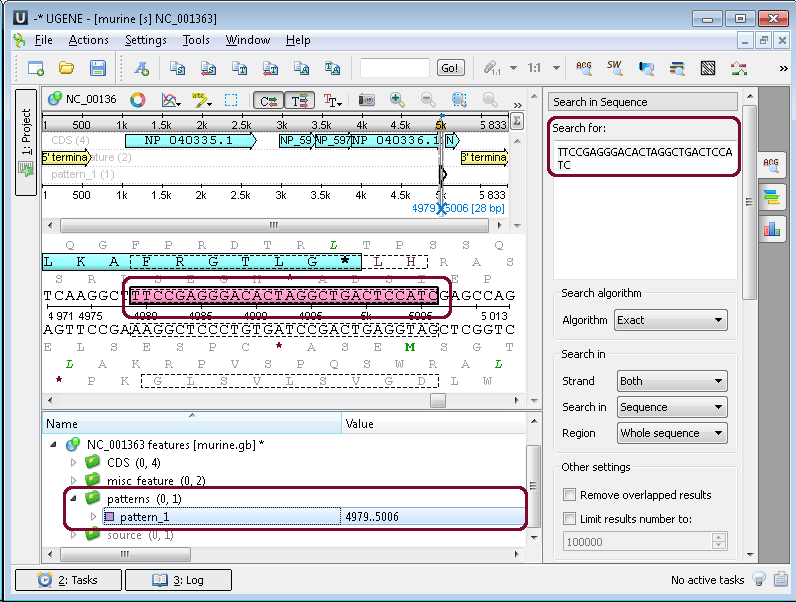
Open the ugene/data/samples/murine.gb by the File–>Open menu, for example. Sequence View with murine.gb opens. Select the Search in Sequence tab of the Options Panel. Click Show more options, and more options appear. Insert, for example, “TTCCGAGGGACACTAGGCTGACTCCATC” pattern into the Search for: field and choose annotation parameters. For example, as in the picture below:
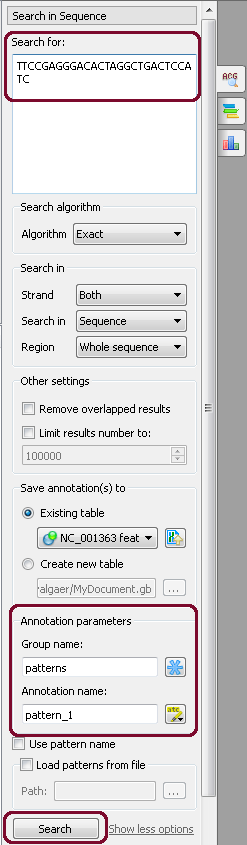
After that, click the Search button. If the pattern is present in the sequence, it appears as new annotation(s):