UGENE Mini-Tutorials: Consensus Sequence, Filter Sequences by Phred Quality Score and More
Generating DNA Sequence
If you need to get a set of sequences, for example, for testing your genome analysis algorithm, UGENE will help you with it.
Just run UGENE via the command-line interface:
ugene –task=generate-dna –count= –length= –out=
In addition, you can set other options of the task, e.g. nucleotide content of sequences or output file format for your genome analysis. Open the help for getting the whole list of options:
ugene –help=generate-dna
or find them in the corresponding documentation page.
Save Consensus
How to save a consensus sequence from a multiple sequencing alignment via the command line?
UGENE provides a ready-to-use solution for extracting consensus sequences from multiple sequencing alignments via the command line. You just need to run the following command:
ugene –task=extract_consensus_sequence –in= –out=
This command extracts a consensus sequence and save it into the specified file. You can use the additional parameters of this command. For example, you can choose the output file format (e.g. –format=genbank) or set the percent threshold of the consensus extracting algorithm (e.g. –threshhold=90).
Here you can find more information about command line capabilities of UGENE.
Filter by Phred Quality
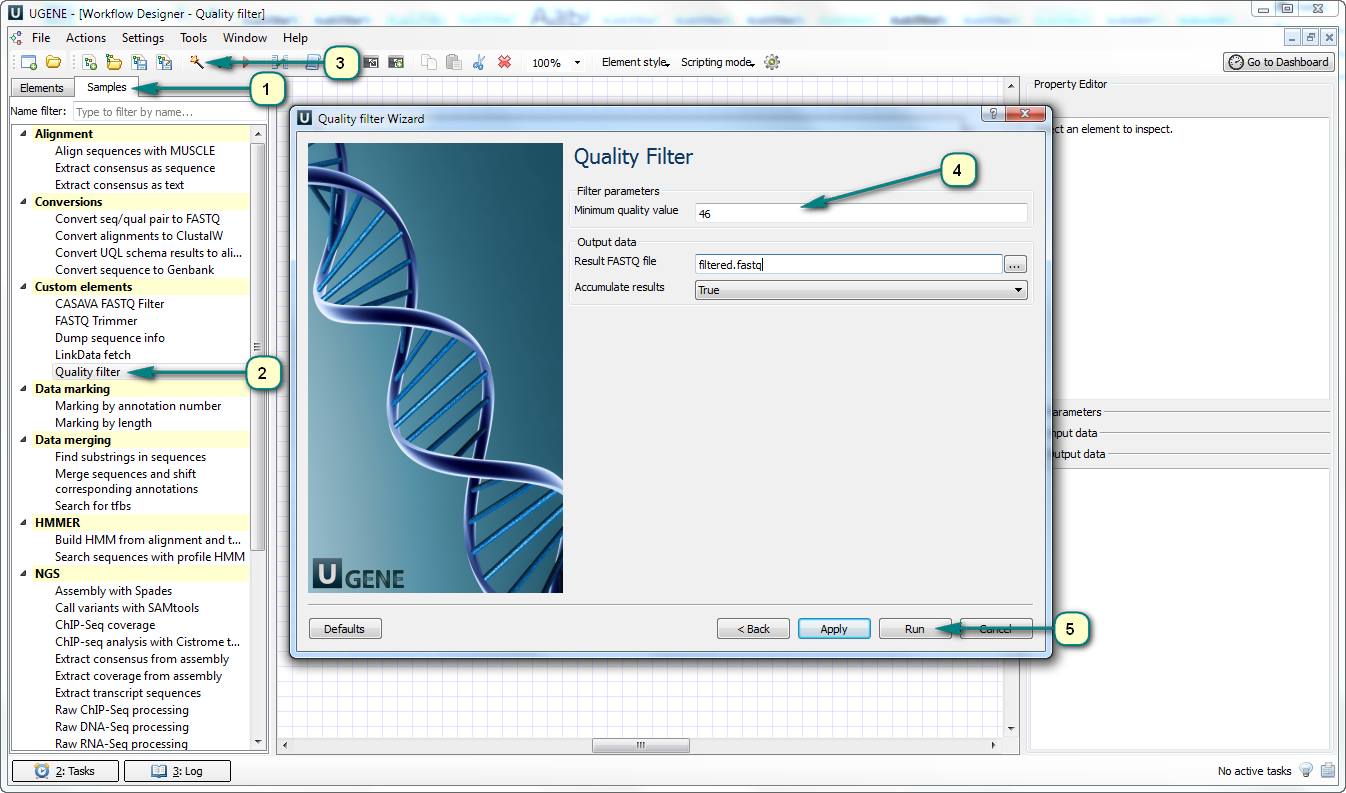
How to filter sequences by phred quality score?
If you want to filter a set of sequences by the quality of sequencing, you can use the predefined “Quality filter” sample of UGENE Workflow Designer.
This workflow has the “Minimum quality value” parameter (threshold). If the quality of one of sequence nucleotides is lower than the threshold then the sequence will be cut off the result file.
Follow the steps for running the workflow:
1. Open UGENE Workflow Designer.
2. Choose the “Samples” tab and double click the “Quality filter” sample in the “Custom elements” category.
3. Click the “Show wizard” toolbar button.
4. Follow the wizard’s pages to set up input files and the phred quality score threshold value.
5. Run the workflow.
Here you can find the documentation about running workflow sample.

