High Performance Computing in UGENE
One of the UGENE main goals is to provide the users with maximum possible performance of their computational tasks. In order to achieve this goal UGENE focuses on integration of highly optimized versions of the most popular bioinformatics algorithms (Smith Waterman, HMMER, MUSCLE etc). UGENE team members have background in bioinformatics and computer science and are also capable to provide custom optimizations to the algorithms.
On this page you can find information about various algorithmic optimizations applied in UGENE.
Smith-Waterman algorithm
UGENE includes several optimized versions of the Smith-Waterman algorithm:
- parallel implementation optimized for multicore systems;
- SSE-optimized;
- GPU-optimized with NVIDIA CUDA;
- GPU-optimized with OpenCL.
Below you can find comparison of various implementations of Smith-Waterman algorithm.
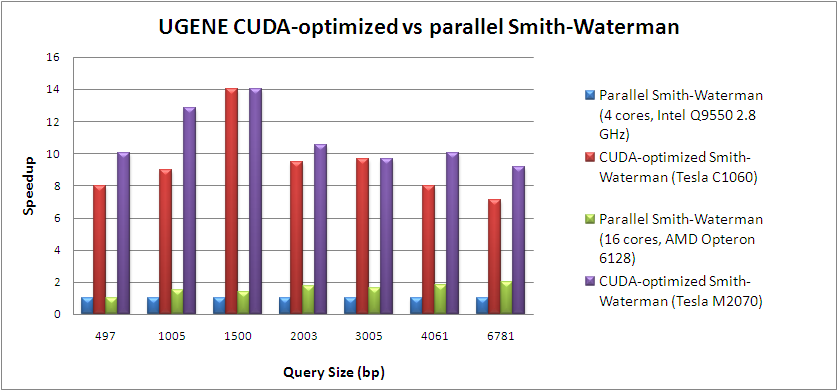
Benchmark data can be downloaded from here.
UGENE Genome Aligner
UGENE Genome Aligner is an efficient GPU-accelerated tool for fast short DNA reads alignment to reference genome. It is rather similar to such popular tools as Bowtie, however instead of Burrows-Wheeler transform it uses rather simple alignment algorithm based on suffix arrays.
We tested the accuracy and performance of the Genome Aligner against Bowtie (Table 1). In this test we aligned approximately 2 million randomly selected short reads with size of 76 bp (experiment ERR008366 from NCBI Sequence Read Archive) to the mouse genome chromosome 1 (GenbankID:AC157543.8).
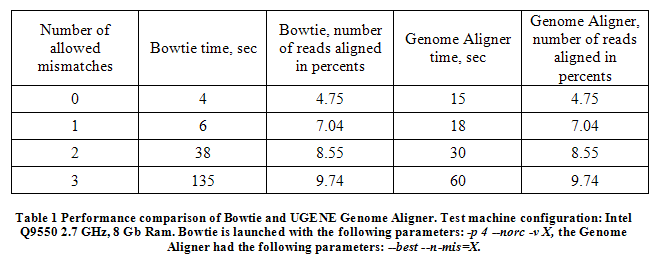
In result a performance boost was demonstrated by the Genome Aligner in the case of 2 and 3 mismatches. Both tools demonstrated equal accuracy.
You can find more details on the Genome Aligner in a draft paper available from here.

