UGENE Tips: Amino-translations, Identical Sequences and more
Amino-translation

Did you know how to get quickly a multiple amino-translation of DNA alignment in UGENE?
To do this open a needed file with a multiple DNA alignment by UGENE. Then open a right-click menu and select { Export -> Amino translation… }. After that a dialog appears in which you can specify a path to the file with the translation, its format and a genetic code that is to be used for the translation. By default the new alignment is immediately opened by UGENE, then you can align it by any of embedded algorithms, for example, CLUSTAL alignment algorithm.
On attached screenshots you can see an example of how to make an amino-translation of HIV-1 strain genome sequences.
See more about amino-translation of multiple alignments in our documentation.
Make a Multiple Sequence Alignment
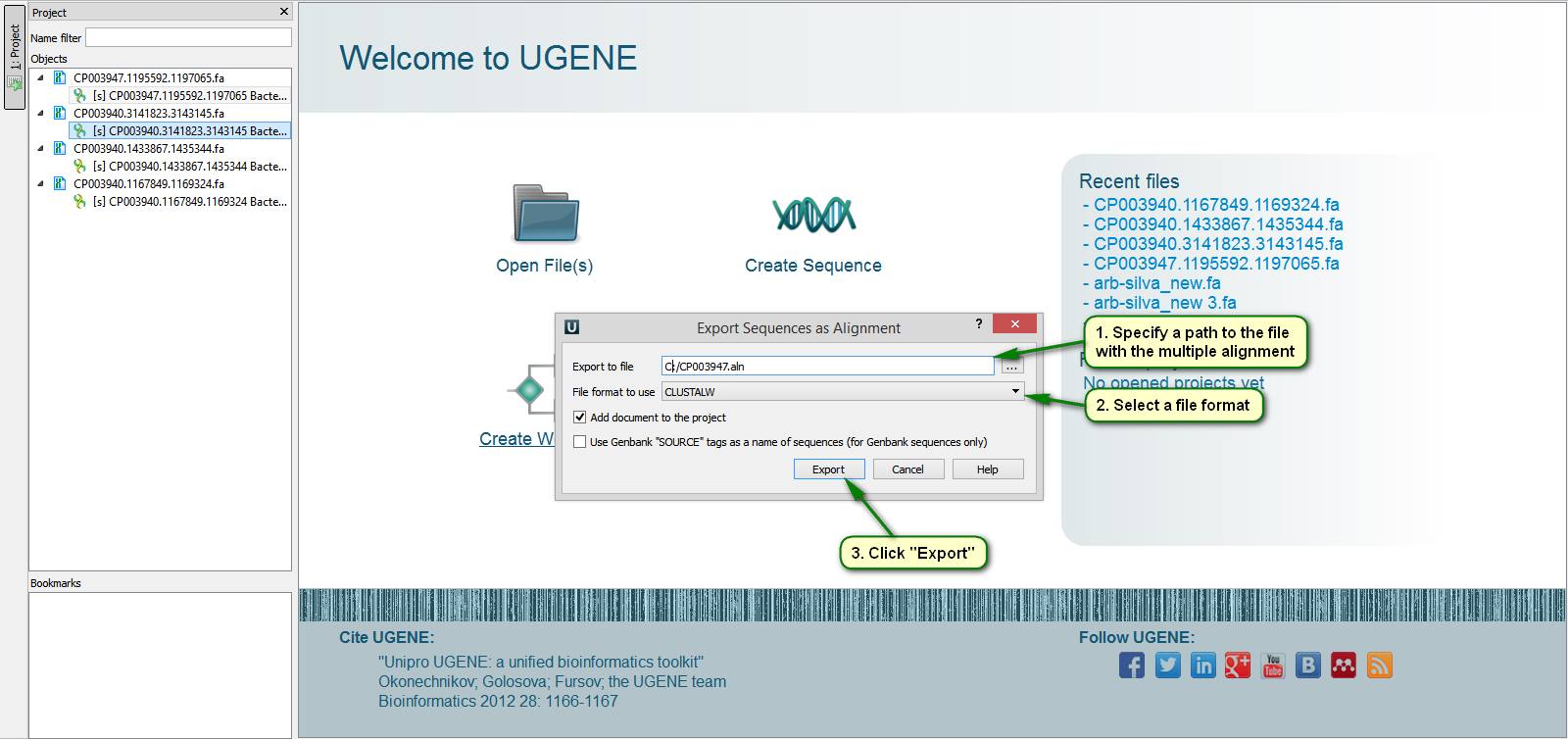
Did you know how to make a multiple alignment of your sequences in UGENE?
Note that UGENE can be a multiple alignment tool and a pairwise alignment tool as well. To do this open your files with sequences in UGENE. Then in the project view select sequence objects that are to be joined into an alignment. Call right-click menu on any of the selected objects and select { Export/Import -> Export sequences as alignment… }. After that in the appeared dialog specify a path to the new file with the alignment and click "Export". By default the created alignment is immediately opened by UGENE. From now on new sequence objects can be added to the alignment using drag&drop from the Project View.
On attached screenshots you can see an example of how to make a multiple alignment of a couple of Cyanobacteria genome sequences.
See more about conversions from multiple alignments to separated sequences and vice versa in our documentation.
Find Identical Sequences
Did you know how to find quickly identical sequences in UGENE sequence alignment viewer?
To do this open your alignment in UGENE alignment viewer and press "Switch on/off collapsing" on the main tool bar. After this identical rows of your alignment with gaps taken into account will be joined into groups. And if you add or remove characters from a sequence constituting a group analogical changes will be applied to other sequences from the same group, i.e. characters will be added or removed from same positions. Thus, the collapsing mode allows editing identical sequences synchronously. Also you can collapse such a group and it will be displayed as a single sequence and all its changes will be propagated to inner sequences.
On attached screenshots you can see an example of how to form groups of sequences and work with them.
See more about grouping sequences in our documentation.