Tutorial: How to Perform BLAST Search in UGENE to BLAST Two Sequences or More
Performing BLAST search and constructing custom BLAST database is available from UGENE with the External Tool Support plugin. In the Settings→Preferences→External Tool Support menu there are two new parameters for selecting the blastall and formatdb executables paths.
Prepare BLAST Database
Now let's activate the „Tools→BLAST→FormatDB“ program menu item. Here one can select the sequences files to create database with, the type of files, the directory to store the DB and the base name for BLAST files along with the title for database files.
Let's create a DB from all the FASTA files from this directory (which is determined by the filter). It is required to specify that the type of the included files is nucleotide. And here's the output files path (same as the input one), the files base name (FASTA_DB) and the DB title (fasta_db).
Please, press "format" and the task is done. Here's the five created database files with the common base name. And, of course, we can perform the BLAST algorithm search. Let's, for instance, open a sequence that had been used to format the DB. And select its region to search in. Now we right-click to bring out the context menu and select „Analyse→Query with BLAST“. The required general options are the database path (this one), and the base name of the BLAST DB files, which is „FASTA_DB“ for the created database. Now that these options are specified, the „Search“ button is available.
We will also specify the annotations group name. The remained options are common BLAST software search options, such as the search type, the e-value, the maximum hits number and the advance search details parameters. Also it is possible to specify how many CPUs you would like to use for the search. We will use all availalbe cores.
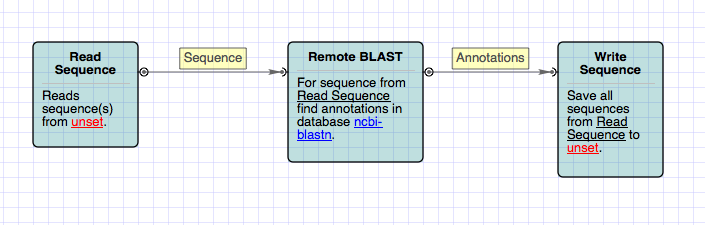
Running BLAST
Press „Start“ and the „Run NCBI BlastAll“ task is being performed. The task is finished, and we can browse the search results. To use BLAST algorithm in a workflow, use the „Local BLAST search“ worker. The sequence to search in goes to the workers input slot, the search results are the annotations of the output Genbank file. The BLAST database to use is specified with the workers „Database name“ and „Database path“ parameters.

