UGENE Tips: Bookmarks, Public Databases and more
Tabbed Documents
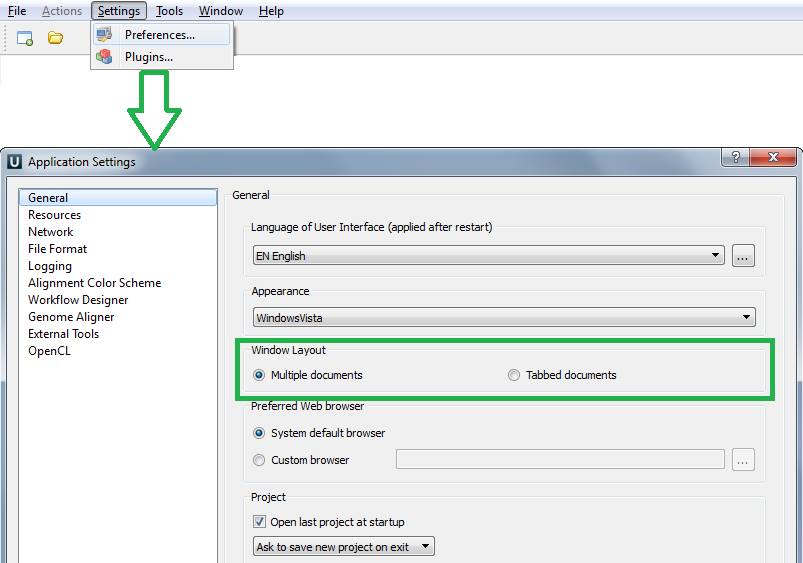
By default different documents are opened by UGENE in separate windows which can be switched by "Ctrl+Tab" shortcut or the "Window" menu on the main menu bar. But those, who are used to working with a few documents at the same time, can find tabbed document view more convenient. You can change the window layout using menu "Settings -> Preferences", then toggle the "Window Layout" parameter from "Multiple documents" to "Tabbed documents" and save your changes by pressing "OK".
The process of setting and the tab mode are depicted on the screenshots below.
Bookmarks
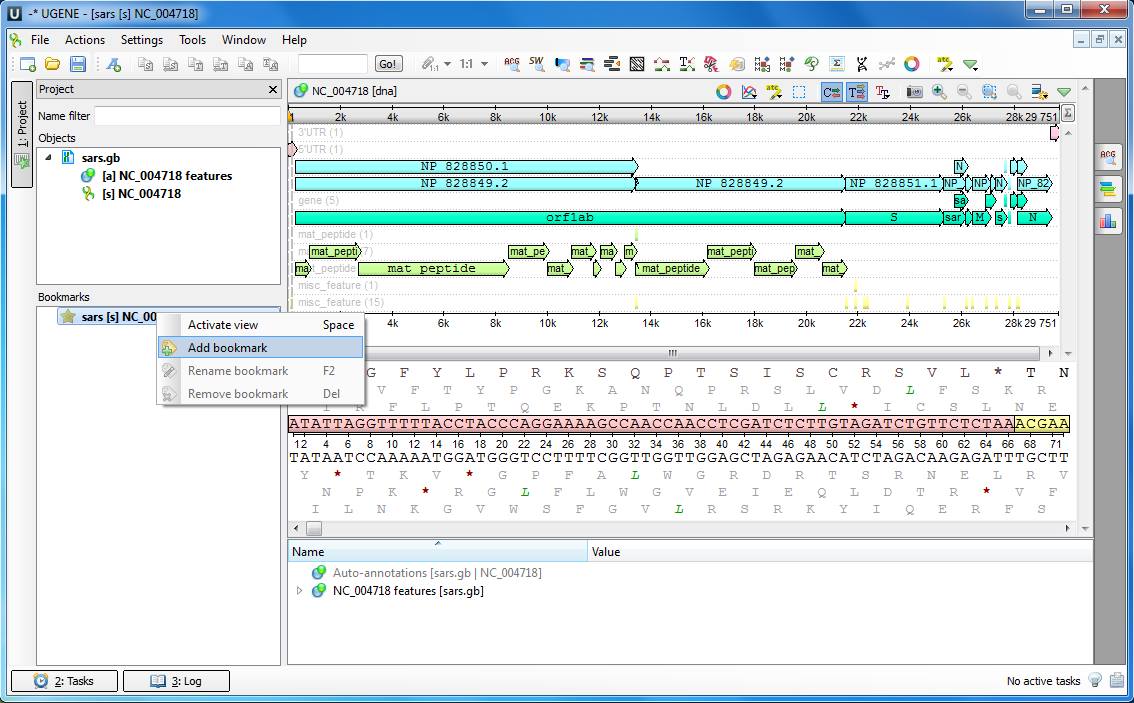
When working with different data types (sequence editing and multiple alignment editing) it is often needed to switch quickly between separate areas of a sequence, a multiple alignment, a genome assembly an so on. For this purpose UGENE provides the Bookmark mechanism. You can navigate to a needed sequence region, for example, and create a bookmark. Afterwards the region will show up immediately upon clicking the bookmark. The list of available bookmarks is displayed in the left-bottom corner of the UGENE main window. All created bookmarks are saved automatically in the current UGENE project and remain available in other sequence editing user sessions.
Public Databases
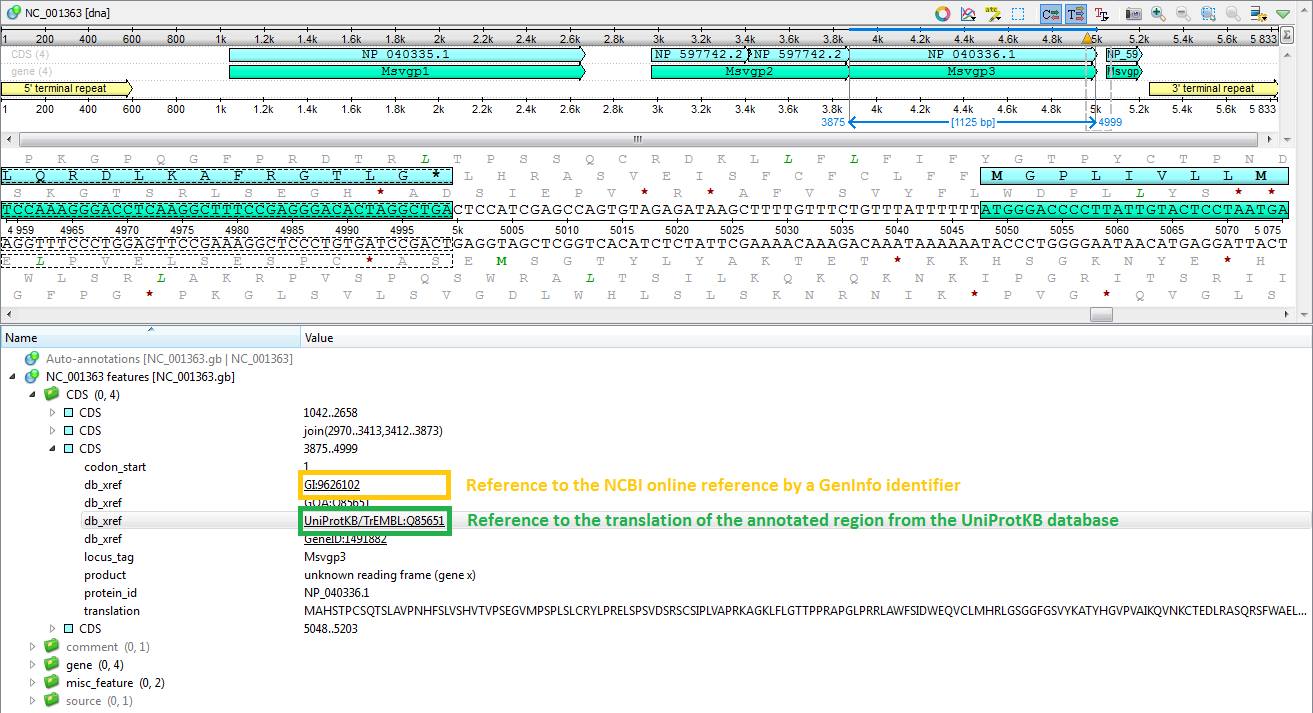
One of the main advantages, when working with sequences from public databases (PDB database, SWISS PROE database, etc.), is the presense of links to related journal articles and reference collections. UGENE can recognize links to external resources, which belong to db_xref category (http://www.insdc.org/db_xref.html), and provide convenient access to them. For example, if a nucleotide sequence annotation contains a link to its translation from some protein database, UGENE downloads and displays an appropriate peptide sequence along with all its annotations upon a mouse click on the link.
Also sequences can comprise links to online references provided by NCBI, EMBL, PDB database, SWISS PROE database, etc. UGENE opens corresponding page in your web-browser, when such link is clicked.
Codon Table
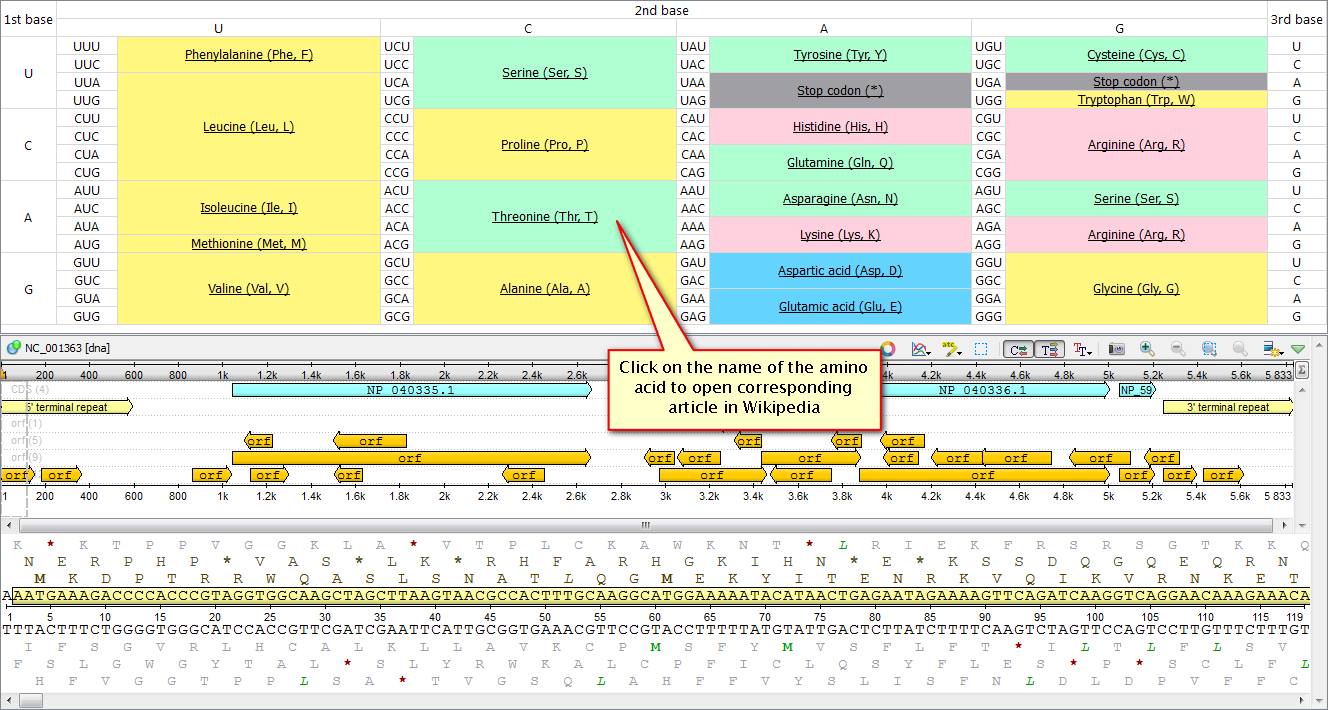
Did you know that there is no need to keep a codon table on hand when working with UGENE? You can easily open it just inside the application. To do this click "Amino translation" on the sequence view tool bar and choose "Show codon table" in an emerged pop-down menu. The table also helps you translate protein back to nucleotide sequence.
If you click on a name of any amino acid in the table UGENE opens a Wikipedia article, corresponding to it, in your web-browser.
Consensus Overview
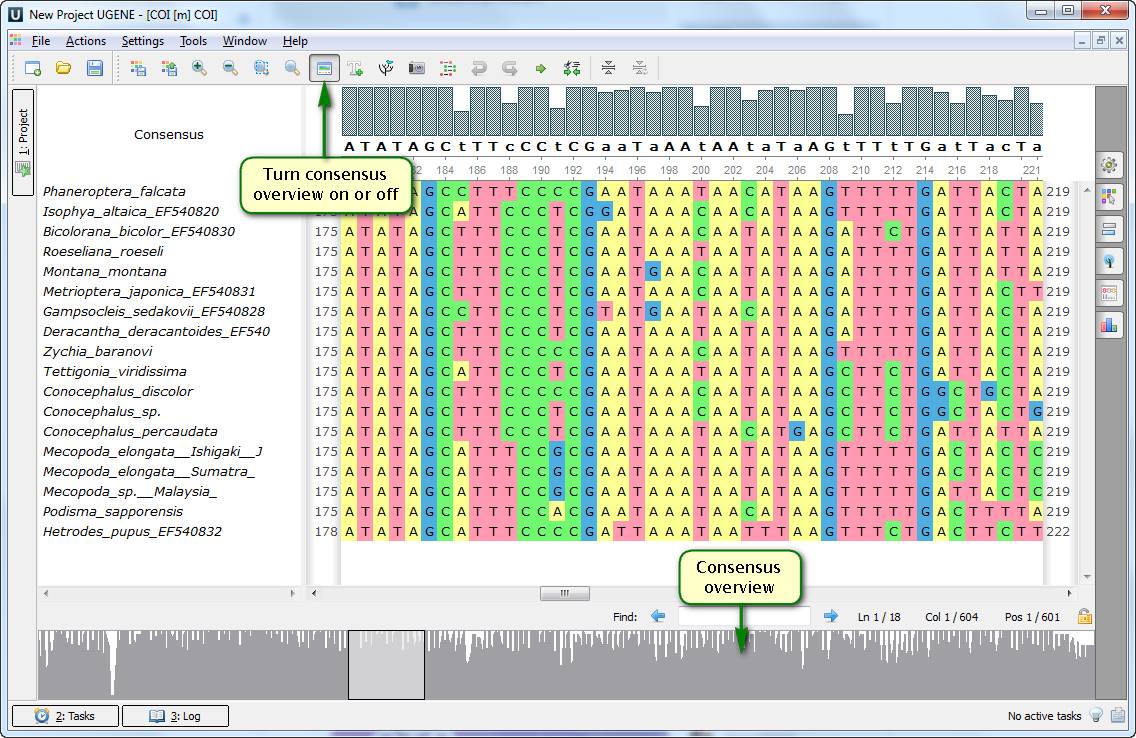
You've probably noticed that since 1.14 version UGENE multiple alignment editor has comprised a consensus overview plot. It allows switching quickly between different areas of an alignment upon a mouse click on a corresponding area of the plot. Moreover, there is an additional option, disabled by default – a whole alignment overview. It enables the quick navigation as well. However, this representation can appear to be more illustrative when you apply custom settings of alignment highlighting to emphasize some regions of it.
You can find more details about the consensus overview in our video tutorial

