Tutorial: Export Multiple Alignment Consensus
This tutorial is about exporting of the consensus sequence from multiple alignments.
General Information
A consensus sequence in a global sequence alignment is just a representation of the most frequent bases in a nucleotide alignment or in a multiple protein alignment. For instance if you have an A letter in 10th position in the most of the sequences in the alignment you will have the A letter in 10th position of the consensus. There are thresholds that you might want to adjust to resolve collisions. For example, in case you have an equal number of letter in a column.
The consensus sequence of an amino acid sequence alignment or a nucleotide alignment is calculated automatically in the UGENE Alignment Viewer. You can see the consensus sequence on the top of the alignment. The number of letter of a consensus each consensus letter is represented as a histogram on the top.
There are several consensus algorithms available:
- ClustalW
- Strict
- Levitsky
- Default Jalview-like algorithm.
Basically those algorithms are equal except different attitude to arguable bases (equal number of letters in a column, inconsistent content). To set the consensus calculation algorithm check the General tab of the Options Panel on the right side of the Multiple Alignment Editor.
Although you can see the consensus in the Alignment Editor, there are several way to get the consensus sequence in a sequence format for further analysis.
The consensus sequence can be exported from UGENE. The first way is to copy a consensus to the clipboard. Find the corresponding item in the consensus context menu.
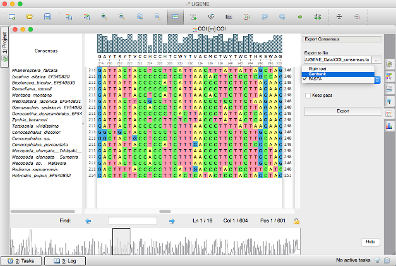
Another way is to use the “Export consensus” tab in the Options Panel. You can choose the destination file and other options of exporting.
Consensus Output Format
Note that the output file format depends on the consensus algorithm. For example, the ClustalW consensus can not be exported into a FASTA file because it is not a DNA, RNA or protein sequence (for an amino acid sequence alignment). There * or N symbols that are bioinformatics notations rather than biological objects. Actually UGENE checks if a consensus sequence of a global sequence alignment can be exported in a FASTA or Genbank format if there are no "bad" characters (pure nucleotide alignment) in the sequence you can see available formats in the File format combobox in Export Consensus tab of the Options Panel. If the sequence cannot be exported into a biological file format because of those characters it will be exported to a .txt file by default.
Export Consensus Pipeline
The export consensus functionality is available from the UGENE Workflow Designer. It might help you to do batch exporting. If you have a number of alignment you want to export consensus from and you do not want to perform repetitive actions. There is the ready-to-use sample. You can use a wizard to set up the workflow in a handy way. Choose “Don’t accumulate objects” if you want to save each consensus sequence in a different file. Results of the workflow are available in the dashboard. Click the file button to open it in UGENE.

