Tutorial: HMMER3 in UGENE
Today we will use a powerful tool for searching sequence databases for homologs of protein sequence and for making a protein alignment, HMMER 3, with UGENE.
The UGENE version of HMMER is aimed to provide a wider user audience with a high-performance HMMER-compatible solution. The plugin with GUI, as well as UGENE itself, is available for all Windows, Linux and Mac OS X users.
The tools are available from „Tools→HMMER3 tools“ global program menu. To search for homologs of a family represented by a multiple protein alignment, we need to build a profile hidden-Markov model, or a profile HMM, for the alignment. Let's open this stockholm alignment as the input file for the build. Now activate „Build profile“ item.
In the opened dialog box we specify the the output profile HMM file for the opened alignment. There's a set of available options. It is possible to select the construction strategy, relative and effective weighting method and adjust e-value calibration. The last tab, „Other“, contains HMMER 3 random generator seed parameter, which also can be specified if needed.
When done with the options, we go back to the the first tab and press „Build“. The profile HMM has been built and saved into the specified location.
Now we can search for the homologs using the created profile. To do so, the sequence to search in must be opened in the project. Let's open this sequence. When the sequence is selected in the project or opened as a view, we can go to the „Tools→HMMER3 tools“ menu again and activate the „Search HMM signals“ item. In the first tab of the opened dialog box we specify the profile HMM file we've built to search with. We will search for the homologically related domains of the opened sequence which are called HMM signals. These signals are represented by annotations and will be stored at the specified annotations table, for instance, a new Genbank file.
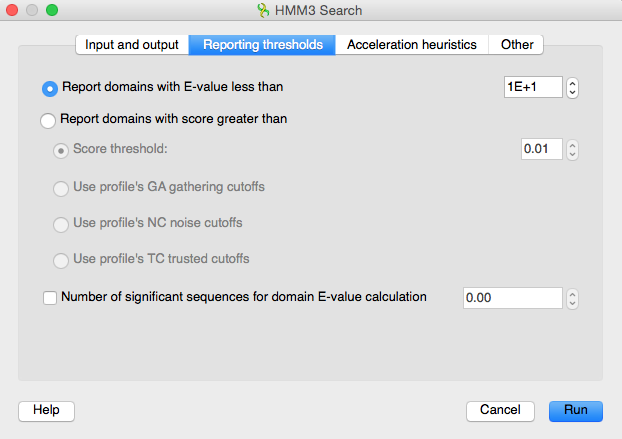
Let's look through the available options. The „Reporting thresholds“ tab contains the filter options. We can search for the domains with E-Value less than the specified number, or for the domains with score greater than this specified number. Also it is possible to search for the domains with score greater than the specified profiles cutoffs parameter value. We will filter the results by E-Value.
The „Acceleration“ tab allows us to accelerate the search if it's needed. When the options customizing is done, we go back to the „Input and output“ tab and press „Run“. The search is done, and we can browse the found signals. The found annotations contain the HMM signals parameters values.
Yet another operation that can be done with UGENE HMMER 3 plugin, besides building a profile HMM or searching HMM signals with a profile HMM, is searching a sequence against a sequence database. The tool is called phmmer. The sequence to search in must be selected in the project view or opened as a view. To invoke the tool, activate „Tools→HMMER3 tools→Phmmer search“ menu item.
In the first tab of the opened dialog box we specify the query sequence file. We will search for homologs of this sequence within the opened sequence. To do so, HMMER will automatically create a special profile for the specified sequence and then use it for the search. It is possible to specify the build options contained at the „Scoring system“ and „E-Value calibration“ tabs. The search options, which are the filter options in „Reporting thresholds“ tab and acceleration options. When adjusting is done, we go back to the „Input and output“ tab and press search. As always, the found results are represented by annotations. We can browse them and read the search results parameters values.
That's how HMMER3 works in UGENE.

