Tutorial: Create a Multiple Sequence Alignment
Here we discuss the hottest topics introduced by our users and show the helpful ways of using UGENE, a free cross-platform genome analysis suite.
Earlier we've been using UGENE MUSCLE multiple alignment tool plugin to create a multiple sequence alignment. We were using a limited number of sequences to align. But would it work the same way with hundreds or thousands of sequences? Today we'll find out.
Open in Multiple Sequence Alignment
Let's open an alignment. This alignment contains, as we see, two hundred twenty three sequences of length up to one thousand three hundred symbols. Let's make another copy of the file to make different alignments of it within one project. To do this, we right-click at the document name from the current Project view and select „Save a copy“. In the appeared dialog box we choose automatically created file name, CLUSTALW file format and we will add the new file to the current project. Click „Save“. The document have been successfully copied and opened at the current project view.
Aligning Sequences
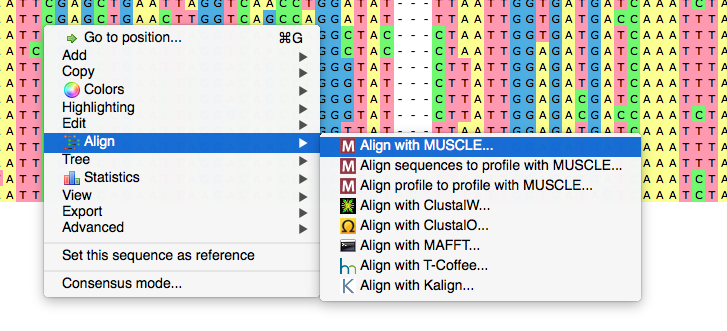
Now we activate context menu by right-click and select „Align“, „Align with MUSCLE“. Let's talk about the MUSCLE sequence alignment software parameters we could customize. First, we can choose a configuration. The default configuration is intended to make the most reliable alignment. „Large alignment“ setting considerably reduces working time at the expense of certain alignment quality variation. The last setting, „Refine only“, leads to „improving“ of the existing alignment instead of making the whole new one. In this case the sequences are being aligned locally at certain parts of the alignment.
We chose the default setting to make the best alignment with MUSCLE multiple alignment tool. Note that you can always check the progress of all active tasks in the Tasks window.
Optionally you can cancel a task. The alignment is done and we can work with it. It consists of sequences of length up to one and a half thousand.
Other Alignment Tools
MUSCLE sequence alignment software works well with alignments consisting of up to one hundred thousand sequences in UGENE. But if we need to work with even more sequences, then we have a special solution, the KAlign tool. Let's apply it to the saved copy of the alignment.
We activate the alignment by double-click on its item within the current project view and select „Activate“. Now we activate the alignment editor context menu and select „Align“, „Align with KAlign“. There are several options here. We could specify gap opening, closing and extending penalties and edit the bonus score. The more the bonus score value is, the more homologically remote sequences will be aligned.
Let's make the alignment with the default settings. It takes only 3 second for task to finish. We can see the resulting alignment. It consists of sequences of length up to three thousand symbols, so it's a different alignment. Also it was calculated four times faster. And it worth mentioning that KAlign is able to deal with alignments containing more than one hundred thousand sequences.

