Tutorial: Practical Tasks of Molecular Biology with UGENE
In this tutorial we will learn practical tasks of molecular biology using UGENE.
Analyzing an unknown virus
This task is devoted to an analysis of an unknown DNA sequence. As an example we will analyze a virus, but in principle this could be any other DNA sequence.
We will go from raw sequencing Sanger reads, then assemble them into a contig and further analyze the virus sequence by BLASTing it against known sequences and creating an phylogenetic tree.
Assembling the virus
We are starting with forward and reverse Sanger reads. (input data NB_forward.ab1 and NB_reverse.ab1)
First we will assemble the reads into a single sequence. Ugene allows to perform assembly of the long sequencing Sanger reads using tool CAP3 assembler.
CAP3 assembler can be accessed from menu Tools->DNA Assembly->Contig Assembly with CAP3
We add both sequences to the assembly and click OK.
The resulting assembly will automatically open in Assembly editor.
Notes:
- There should be an option to open it in MSA editor – for example open the reads in MSA, an visualize the chromatograms
- MSA Editor doesn't allow to export consensus
Next we export the consensus sequence with minimum coverage of 2.
Analyzing the sequence
We further analyze an unknown sequence by running BLAST on it.
Use remote BLAST for this purpose. It can be accessed from menu
Each result represents a sequence. We can download the sequence and create an alignment out of it.
To perform this task select interesting annotations and use context menu item Fetch sequences from annotations by ID
We then convert the file to multiple alignment. Using MSA editor we can perform multiple alignment of the sequences.
Color scheme based on mismatches allows to easily see possible mutations in the virus.
One last step could be building of a phylogenetic tree of the sequence.
PHYLIP or Mr Bayes can be used for this purpose.
Preparing primers for a RT PCR reaction
In this hypothetical task we try to find primers for a particular sequence with UGENE as a primer design program.
Let's assume that we interested in how to design primers for PCR for a mRNA sequence of a well known fusion gene TMPRSS2-ERG which is known to be a marker for several types of prostate cancer.
Searching for the sequence
If we the sequence is not available, we can try searching it a sequence database, such as NCBI Genbank
UGENE allows to search sequences in Genbank directly.
We will use NCBI search interface for this task.
It can be accessed from main menu File->Access NCBI Genbank
We are setting our query to TMPRSS2/ERG. To further specify the search we set the organism as human.
Several results will be available as a table, each raw representing a single result. More details about the sequence are available in the Details field of each raw.
As one can see there are several version of the transcript. One can download all of them and examine the the details.
However for this tutorial we will select one particular sequence (FJ423744) and download it.
How to design primers for PCR
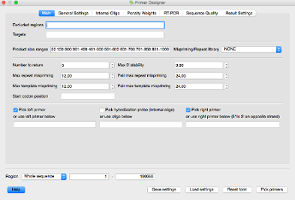
UGENE integrates Primer3 – a popular primer design program.
Let's apply it to design primer marker for our sequence.
One can access Primer3 from menu Analysis -> Find primers…
If you used Primer3 earlier from web-interface you will have no problems with using it in UGENE while the interface is very similar. Each option or field has an interactive hint which explains it.
To start searching for primers just click Pick Primers.
Corresponding primer pairs are shown as annotations. Detailed information about each pair such melting temperature and etc can be found in the annotation features using Annotation Editor.
(Screenshot: annotation editor and primer pair properties)
One tricky part when designing the fusion sequence is too make sure that the left primer lies in the 5' gene of the fusion, while the right primer lies in the 3' gene of the fusion
This can be achieved by excluding the junction region from the primer analysis.

