Tutorial: Search for Sequence Repeats with UGENE
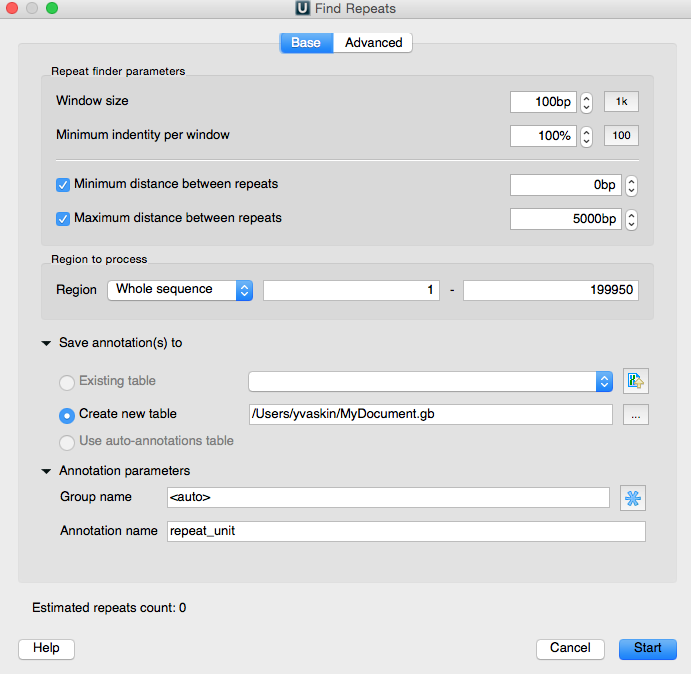
Today we will search for repeats in a DNA sequence in UGENE. This is one of the types of biological sequence analysis that UGENE ca do. To do so, we open such a sequence, for example of the FASTA format. Now we activate the sequence view context menu and activate „Analyze“, „Find repeats“. The „Find repeats“ dialog box appears and provides us with a set of options to specify the search parameters. Let's consider them.
Repeats Search Parameters
First of all, we can specify the minimal length of the sought-for repeats and their identity rate. The identity rate is the percentage of matched-unmatched bases in the potential repeats. We will search for repeats of the 30 b.p. minimal length and with 100% identity.
Further, the minimal and maximum distances between repeats can be set. Also we can choose the algorithm to search with. And it is possible to specify whether or not the nested repeats should be filtered (meaning a repeat in a repeat), and whether or not the algorithm should search for the inverted repeats.
At the panel below we can specify the sequence region to search in. The search results are represented as annotations. So, as usual, we will specify the annotation table to save the results into, and specify the group name and the annotations name.
Additional Options
When ready, we press „Start“. The search is done, and we can see the result annotations at the panoramic view. Let's navigate among the annotations by selecting the desired annotation item in the annotation editor. As we can see, each result annotation consists of two joined regions — the repeats. The results can intersect, but there are no nested results since we didn't check the „Do not filter nested repeats“ option.
Now, let's invoke the repeat finder one more time. 3 more options became available which allow to specify the search relative to the annotated regions. So, we can search repeats that lie inside of an annotated region or have an annotated region inside. Also we can filter the results with annotated regions inside. Let's set the results minimal length to a small value by pressing the heuristic „up to 1k results“ button, and perform search for the repeats that lie inside of the regions annotated with annotations named „repeat_unit“. Press „Start“.
As we can see, we have a repeat, that lie completely inside of the specified annotated region that represents another repeat.

