Tutorial: How to Search for Restriction Enzymes
Here we discuss the hottest topics introduced by our users and show the helpful ways of using UGENE, a free cross-platform genome analysis suite.
Search for Restriction Enzymes
With UGENE restriction site finder you can search for restriction enzymes cut sites in a DNA sequence. Today we will use the restriction site finder plugin to perform such a search. Let's open a DNA sequence, for example, another Influenza H1N1 DNA researching result. When the download is finished, we see the annotated sequence.
To evoke the plugin, we activate the context menu and select „Analyze“, „Restriction sites…“. Let's consider the available options here.
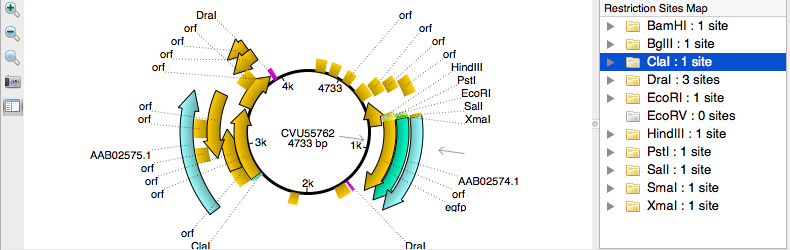
List of Restriction Enzymes
First of all, we can specify the Enzymes file, pressing the corresponding button. UGENE package contain two recent enzyme database files each with the list of restriction enzymes: the full and t2 versions of REBASE database. We choose the full one.
The opened enzymes are represented at this table. Here we can unfold an enzymes group, see the detailed information of the enzymes and select items for restriction enzyme mapping by checking their check-boxes. „Select all“, „Select None“ and „Invert selection“ buttons help to specify the selection, and the „Save selection“ button allows us to save the specified selection for later use. The numbers after a group name is a pair of the currently selected enzymes number and the group capacity. If we press „Select all“, we can see, that all the groups numbers become equivalent. Now we press „Select None“.
If we need more detailed information of the concrete enzyme, we can select the desired enzyme with mouse. Then pressing the „REBASE info“ button will lead us to opening a REBASE web page with detailed information of the enzyme.
Restriction Enzyme Mapping
Let's finally select item to perform restriction enzyme mapping. Note, that the selected enzymes are enumerated in the list of restriction enzymes at the text box located below the selection table.
Further, we can specify the sequence region to search in, and set the results number filter. If the filter is active, then we will only get the results whose number exceeds the minimal value and is less than the maximum value.
Finally, we need to specify an annotation table to save the results into. We will save the results into a new file, and enter „Enzymes“ name for the results group. Click „Run“.
The new document, containing all the results annotations, is added to the annotations editor. And we can see the results annotations sorted by enzymes at the Panoramic view. Let's select some enzyme from the annotations editor. We can see it selected at both panoramic and detailed view. We also see two triangles near it at the detailed view. They represent the enzyme cut sites.

