Tutorial: UGENE as a Tandem Repeat Finder
UGENE tandem repeat finder provides a fast and memory efficient way to search for short tandem repeats. You can use it for pedigree determination or for other purposes like finding variable number tandem repeats.
Preparing Data
I will open a large sequence this time. Namely this hypothetical protein gene of nostoc cyanobacteria of length over 8 million base symbols. When the sequence is loaded, I right click at the sequence view and select „Analyse→Find Tandems“.
Setting Parameters
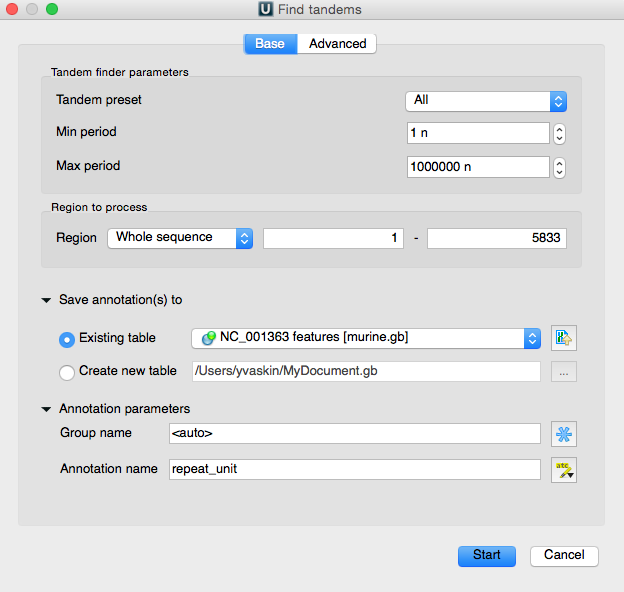
The opened dialog box contains the short tandem repeat search parameters. Simple tandem repeats preset option allows to set such search parameters values that corresponds to particular short tandem repeat groups such as micro-satellites, mini-satellites or big-period tandems. If needed, the parameters can be set manually. We will use the {„micro-satellites“ | „mini-satellites“} preset.
Min and max period parameters corresponds to the minimum and maximum acceptable repeat length measured in base symbols.
The algorithm parameter allows to select the search algorithm. The default and a fast one is optimized suffix array algorithm.
Minimum tandem size sets the limit on minimum acceptable length of the tandem, id est the minimum total repeats length of the sought-for tandem.
Also it is possible to set the minimum repeats count.
The last option, „Show overlapped tandems“, allows to specify whether or not the plugin should search for the overlapped simple tandem repeats.
Running Short Tandem Repeat Finder
After the search options are set, we can specify the region to search in. The search results will be stored and displayed as annotations.
When done, press „Start“.
The variable number tandem repeats search in the loaded sequence is done. We have found over three thousand results.

