Tutorial: UGENE as a Transcription Factor Binding Site Prediction Tool
Today's topic is SITECON plugin, which is a transcription factor binding site prediction tool. The tool is developed in Institute of Cytology and Genetics SB RAS.
SITECON Description
The basis of the search is a set of conservative (conformational and physicochemical) properties of transcription factor binding site alignments. These alignments conservative properties are stored in SITECON profiles (also known as SITECON models) – transcription factor database of SITECON. UGENE package contains pre-built profiles for over 90 sample sites, but our users can also create their own profiles of TBFS alignments.
SITECON Transcription Factor Binding Site Database
To show how to search for transcription factor binding sites of a DNA sequence, I will open a nucleotide sequence human_t1.fa. The next step is to invoke the plugin: right-click at the opened view and select „Analyse→Search TBFS with SITECON“. The opened dialog box contains „File with model“ field. Let's choose the model from the transcription factor database to search with by pressing „Browse“. Here's eukaryotic and prokaryotic profiles groups, which contain pre-built profiles. Let's choose one of them.
When the model is selected, the „threshold“ filter is populated with a set of available thresholds read from the profile. Adjusting the threshold helps to filter low-scoring results. We see the default threshold, and can browse and select others by invoking the drop-down list.
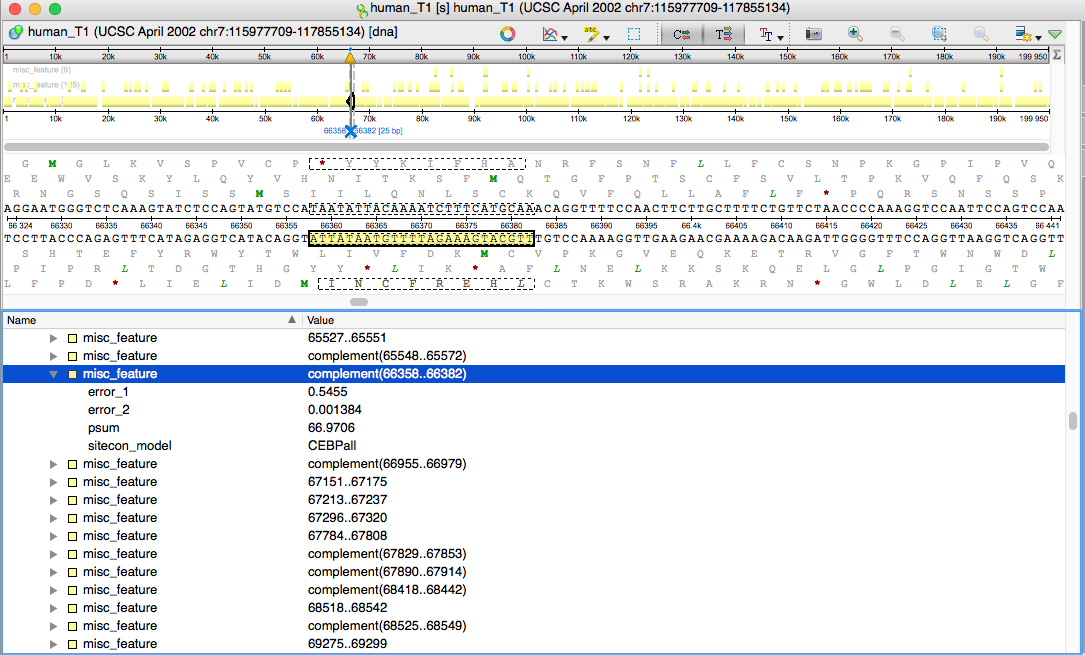
Running Search for Transcription Factor Binding Sites
The options are standard search options: strands and the sequence region to search in. Press „Search“.
The search results are placed into the dialogs table as table rows and contain PSUM and first and second type errors values. To browse the results as sequence annotations, press „Save as annotations…“ and specify the annotations table to contain the resulting annotations.
To create a specific binding site alignment profile for future SITECON searches, activate „Tools→SITECON→Build new SITECON model from alignment“. In the opened dialog box specify your input alignment file location (the alignment must contain no gaps), and output profile location. After that, window size and weight algorithm may be selected. Also calibration options are available to set.
When done with the options, press build. The profile is built and saved, and can be selected as a profile for a TBFS search.
That's how you can search for TBFS with SITECON in UGENE and create specific binding site alignments profiles. Thank you for watching.
Advanced Options
Window is used to pick out the most meaningful alignment region and is located at the center of the alignment. Must be: windows size <= the alignment length, and recommended to be: windows size <= 50 and is size must be not less than the consensus sequence for this particular site and should not exceed transcription factor binding sites alignment length. Optimal is to choose this parameter a bit less than the alignment length, but not more than 50.

