UGENE 1.14 List of Features
Here we discuss the hottest topics introduced by our users and show some helpful tips for using UGENE, a free crossplatform genome analysis suite.
In this tutorial, we will show you a brief overview of the new features of UGENE 1.14.
The new useful thing is the help system embedded into UGENE. In each dialog you will now find a Help button. Click this button, if you want to read the corresponding online documentation about the dialog.
The new documentation is based on the Atlassian Confluence system that makes the documentation more useful than the previous version. For example, you can search for the required information using the system.
Plasmids and Views
It is now possible to mark a sequence in UGENE as circular for visualization of plasmid maps. The sequence must be stored in the GenBank format, if you want this mark to be saved between different UGENE sessions.
Making a sequence circular affects how a pattern is searched in the sequence. So, if the sequence is circular, results located over the end-start junction of it are also found.
Another feature for plasmid maps is auto annotation of the plasmid features like promoters, regulatory regions, primers, and so on.
Multiple Sequence Alignment Editor
It provides the multi sequence alignment overview now. This feature is useful for watching the whole picture of a multi sequence alignment and fast navigating to interesting regions. You can adjust the overview picture in both ways: viewed information and look and feel such as graph type, orientation and color.
The consensus sequence of your alignment can now be exported. Select the corresponding options panel tab to do the export. Note that the consensus type can be selected on the General tab of the panel.
Phylogenetic Trees
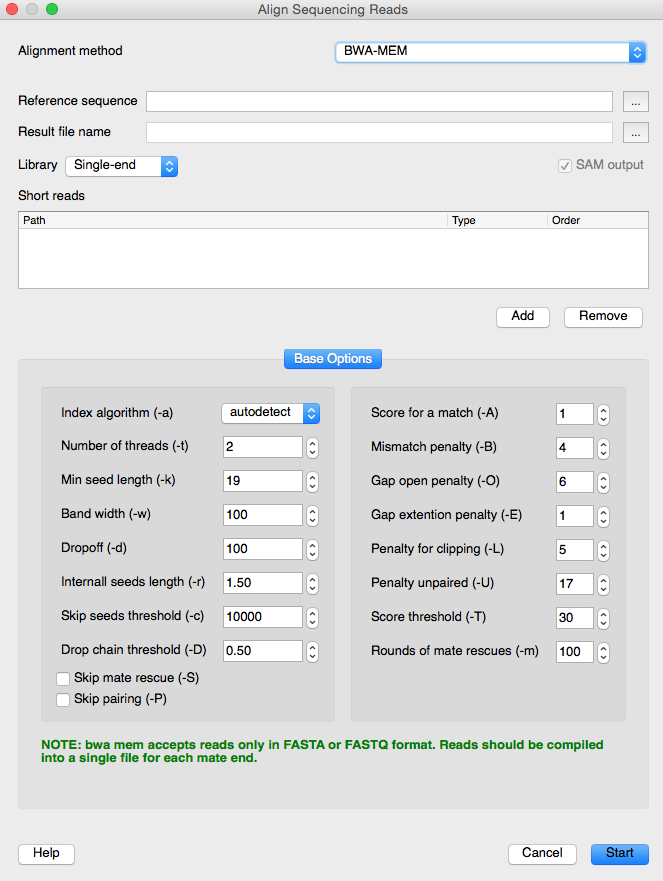
Besides Bayesian inference and Neighbor-joining methods there is a new algorithm for phylogenetic tree building in UGENE. The BWA-MEM program has been integrated into UGENE as an external tool. The program provides building phylogenetic trees using the maximum likelihood algorithm.
There is a new feature of the tree viewer. Right click on a node and choose the “Reroot” item to change the root of the tree. UGENE will recalculate the distance scores automatically.
NGS
The NGS data analysis area has also been improved in UGENE. The BWA-MEM tool is available from the “Align Sequencing Reads” dialog.
Also, there are several new utility elements in the Workflow Designer for working with NGS data. The elements helps with executing different action on BAM files such as sorting, merging and filtering; FASTQ files such as CASAVA trimmer and quality trimming; and others.
Shared Storage
Last, but not least, UGENE provides a shared storage feature. Using it, you can easily share your bioinformatics data with your lab colleagues. Try this feature with the UGENE public storage that is available for free. See the details in another video.

