UGENE Tips: Multiple Sequence Alignment Colores, Dot Plots and more
Multiple Alignment Highlighting
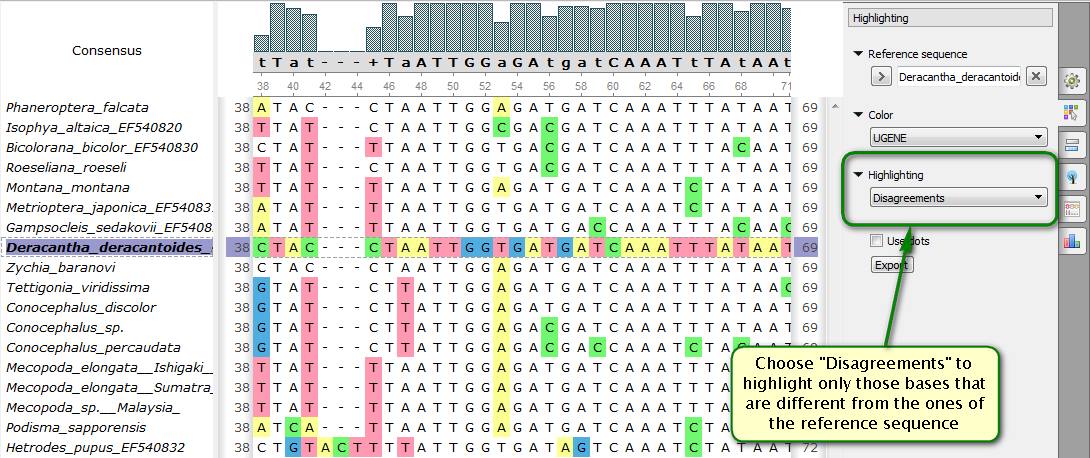
Did you know how to make a multiple alignment more illustrative with UGENE? It's often needed to evaluate similarity or difference between one sequence and the others. Specially for this purpose the UGENE Multiple Alignment Editor allows specifying a reference sequence that the others are compared with. The difference is shown using different highlighting modes. There are two basic options available:
1. Color schema.
2. Highlighting area.
Particularly, you can make UGENE highlight only those bases that are different from those of the reference sequence in same positions. Or vice versa, only bases that are the same.
As for color schemes, for example, UGENE can color all the bases depending on their similarity to the reference sequence in the current position. Bright colors correspond to different bases, the dark ones are to same bases. Color palette can also be grayscale that is very useful if you add alignment image to your publication.
You can learn more information about highlighting in the UGENE Alignment Editor in our documentation.
Multiple Alignment Coloring
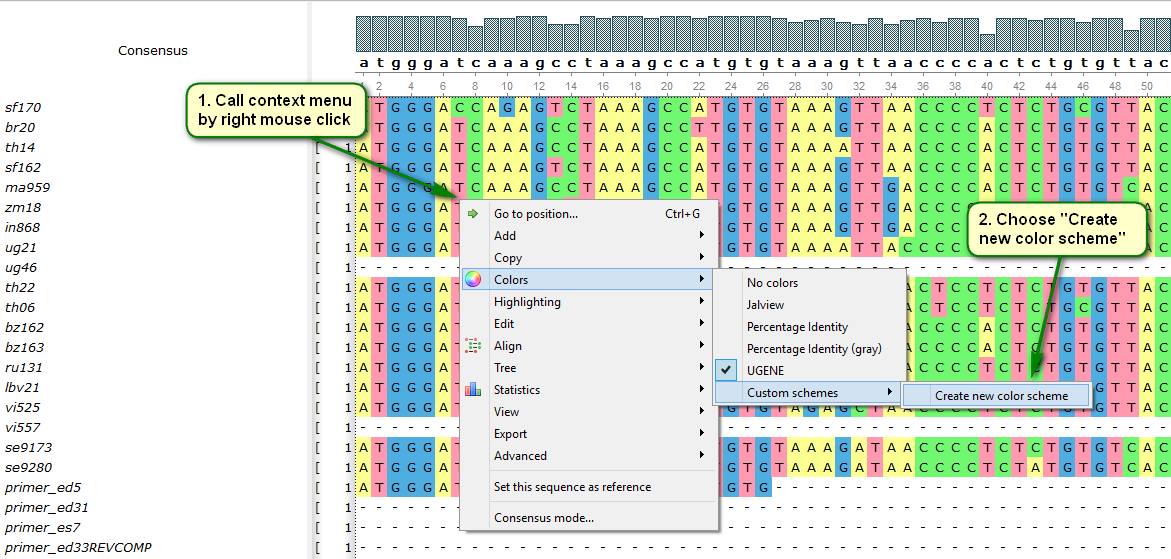
Did you know that literally any color can be used for multiple alignment coloring in UGENE? This is achieved by custom color schemes. Basically, a color scheme is a correspondence between some alphabet (nucleotide or protein) characters and colors that are used for coloring appropriate bases if the scheme is applied. Being once created, color schemes are saved permanently and can be used at any time.
The new color scheme creation process is depicted on attached screenshots.
In order to apply a present custom color scheme to a multiple alignment choose the following items in the alignment editor context menu: Colors -> Custom Schemes.
You can learn more information about custom color schemes in the UGENE Alignment Editor in our documentation.
Building Dot Plots
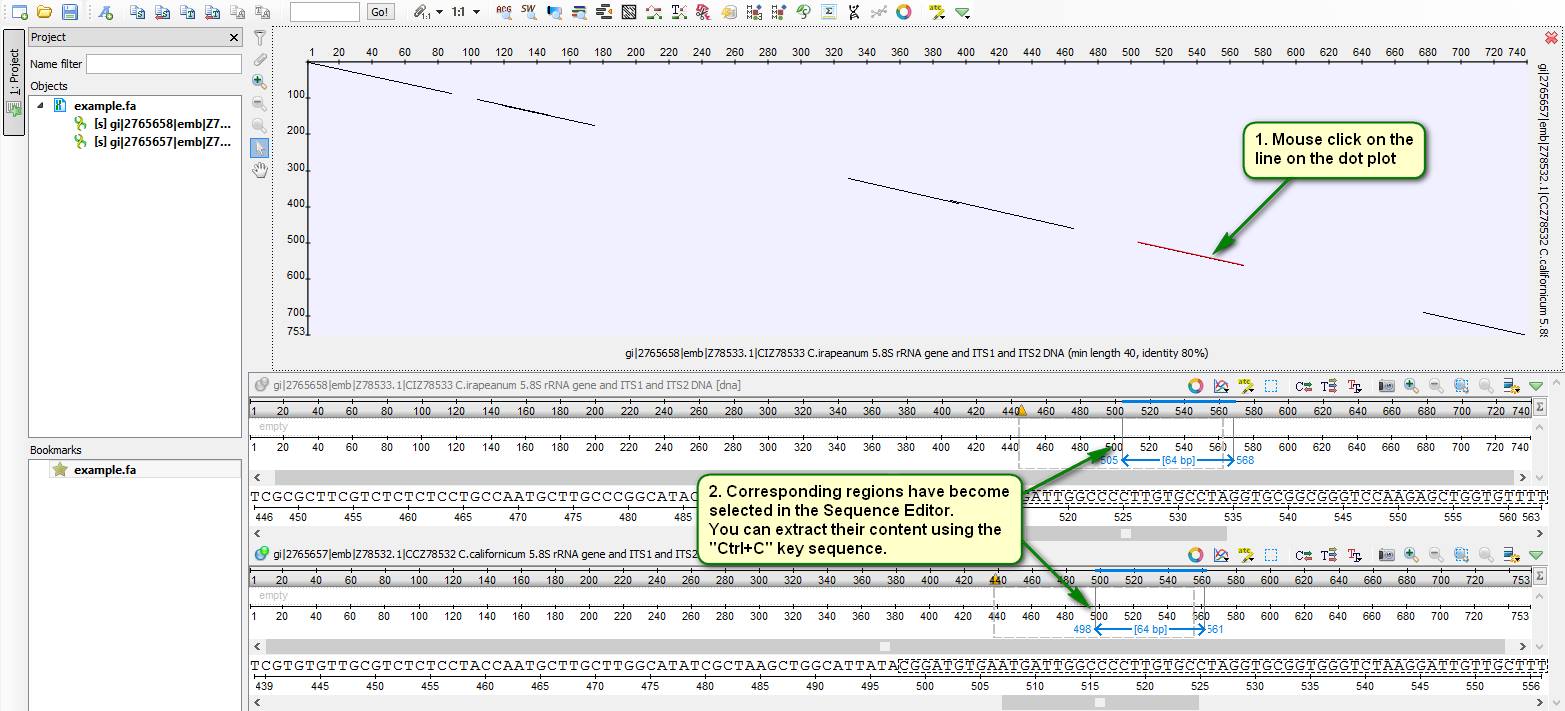
Did you know how to assess quickly a similarity between two sequences?
The UGENE Sequence Editor allows building a dot plot for two given sequences that shows clearly mutual regions having required similarity.
The dot plot building procedure is depicted on attached screenshots.
See more about the UGENE dot plot capabilities in our documentation.
Access Remote Database
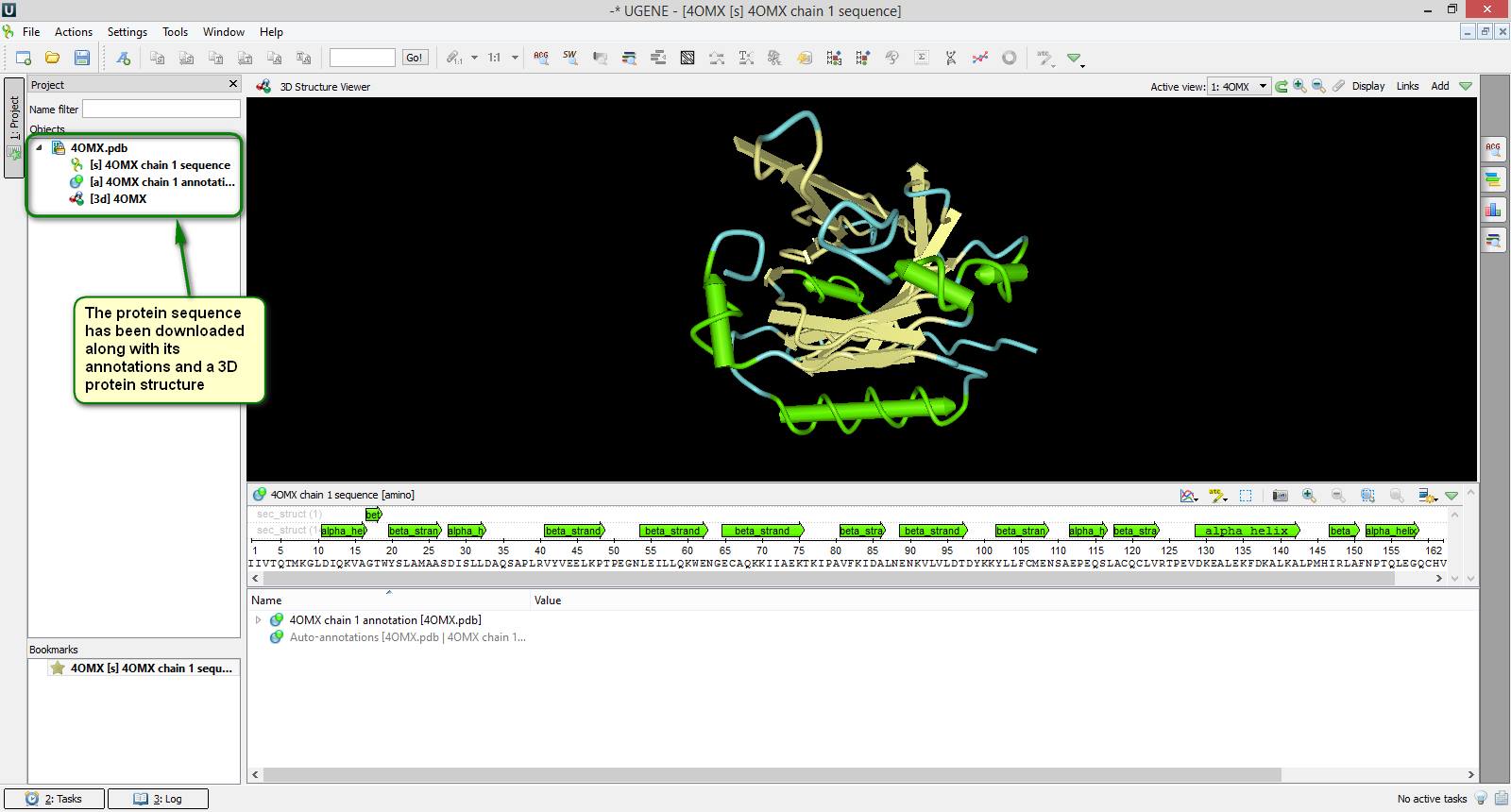
Did you know that you can access many known public databases (e.g. TREMBL) just from UGENE?
To do this choose the { File -> Access remote database… } item in the main application menu. Then you should specify the needed database and the accession number of your sequence in the appeared dialog. Confirm the input parameters by clicking "OK" and the sequence will be opened in UGENE in a few moments. Note that you can access multiple sequences at once. Just type identifiers of the sequences separated by a space or semicolon and they all will be loaded in UGENE.
You can see an example of how to download a sequence from the PDB database on attached screenshots.
See more about working with public databases in our documentation .

