Tutorial: UGENE Multiple Alignment Overview
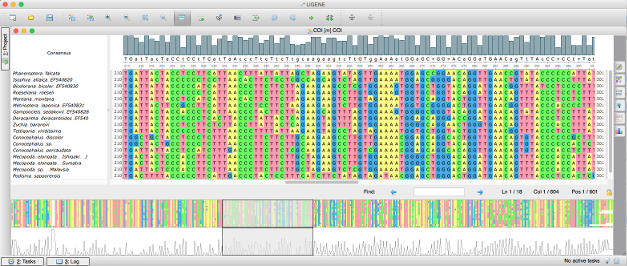
Multiple Alignment Editor has many features common to multiple sequence alignment tools like highlighting of diffidences to spot mutations, finding a subsequence in an alignment and gap removing. But sometimes you want to see the alignment as a whole, that is where the overview might help, this is one of the features that you do not usually find in other multiple sequence alignment tools.
General Overview Information
This tutorial will show you how to use the overview of the UGENE Alignment Editor. The overview is the quite powerful feature because it helps to make many things simpler and easy to spot. Note that the histogram overview feature is shared across UGENE components. There is the same coverage look-and-feel in the UGENE Assembly Browser.
To see the overview open a file generated by an alignment program with UGENE and take a look at the bottom part of the alignment window. It can be opened for a nucleotide sequence alignment and a protein sequence alignment as well.
By default, the overview repeats the consensus graph on the top of the window, but it shows the whole view of the graph. Using the overview, you can see the regions of an alignment, where sequences are different, and easily navigate to these regions. Also you can easily spot low-coverage or high-coverage regions. If a mutation goes into a low-covered region it might mean that we have a false mutation.
Using the overview you can navigate to any part of your alignment. Just click on an overview region or drag the sliding window to move across alignment in the main window. Essentially, the overview and the main alignment window are synchronized. So when you change characters in the main window or move it, the changes are passed to the overview as well.
Overview Options
It is possible to change the information shown by the overview. For example, you can choose the gaps overview mode to see the regions of an alignment that contain gaps. By default the histogram overview is shown. It visualizes your alignment data as a coverage graph. Also it is possible to show "Simple" overview which is just a bird-eye view your alignment with a selected color-scheme. using the "Simple" overview you can spot alignment-wide mutations for a and changes for a protein sequence alignment.
The simple overview is helpful in combination with different highlighting modes of the Alignment Editor. For example, set the reference sequence of your alignment and choose the disagreements highlighting mode. It provides a way of fast and easy observing of all disagreements in the alignment. The overview will be changed as you change your alignment highlighting scheme. The same is true for the "Simple" overview.
The look and feel of the graph overview can be adjusted. It is possible to change the graph type, color and even orientation. When you change the orientation the graph is just flipped upside down.
The adjusted picture of the overview can be exported to a file. The file format is a standard image format (PNG, JPEG, BMP, etc.)You can choose what type of overview the destination picture should contain: simple, graph or both of them.
Additional Materials
UGENE alignment program documentation page

