Tutorial: UGENE PCR in silico
PCR primer designing is a day to day lab activity. Also you might want to check if a designed primer that your lab uses is applicable to your new sequence. By checking your primer (e.g. melting temperature of primers or primer hairpin) with in silico PCR features you can track some parameters of your PCR experiment before it. It is extremely easy and a cheap way to not get into trouble.
This tutorial is about the In silico PCR primer design feature in UGENE.
PCR Design Tools
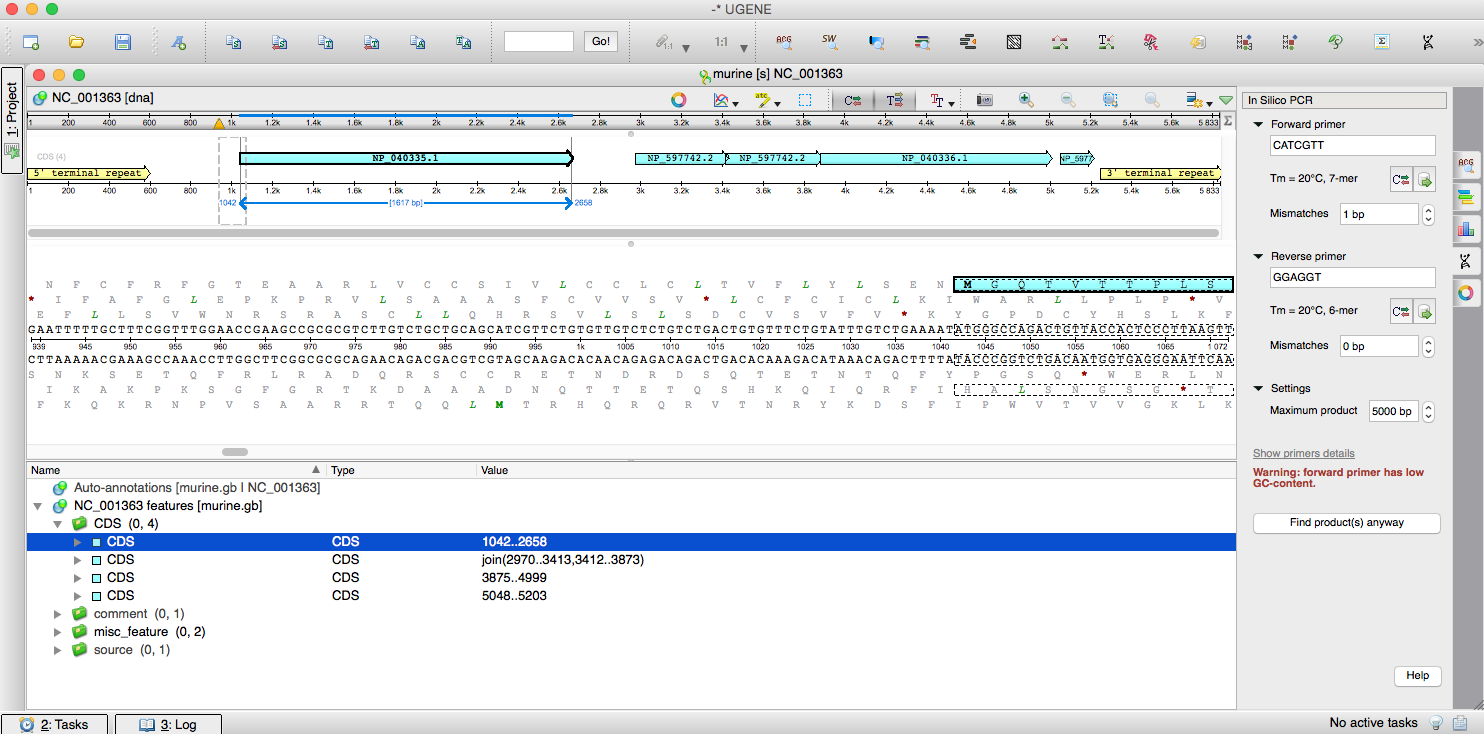
You can run the simulation of your PCR experiment for any nucleotide template. You might want to check primer design tool Primer3. For running In silico PCR open a DNA sequence which contains the target region you want to amplify; and activate the PCR options panel by clicking its icon. You can find this icon on the right side of the UGENE Sequence View.
Enter two primers’ sequences for setting up the simulation. There are two ways to do it: to enter them manually or to find them in the primer library. You can type the sequence or paste it manually. The primer library allows you to reuse your sequences (even across a lab), but you need to populate it in advance.
If you need to reverse a primer, click the corresponding reverse-complement button.
UGENE automatically calculates the parameters of primers. You can see the values in the Primers Details dialog. There are GC content, melting temperature of primers, self- and hetero-dimers (primer hairpin) and others parameters in the dialog.
Primer Checking
If a value of a parameter does not fit into the predefined boundaries then the value is colored with red. It means that you should pay attention to it. Maybe, your primer pair is incompatible for In silico PCR and real PCR in turn and may result it no amplification. In addition, one of the warnings is shown in the options panel.
There is a couple of parameters in the options panel: the maximum size of the target amplicon and the amount of mismatches for binding of each primer. Choose the parameters values and click the “Find products” button. It will just find the primer sequences that you enter in a sequence opened.
After that, you can see the list of probable amplicons – the PCR products. Click a product for seeing its region in the sequence view. Double click the product for exporting it to a file.
Primer Database
Primer designing can be skipped by using old primers. If you have a set of pimers that you usually use you can add them into the UGENE primer library. It will simplify your work with In silico PCR primer design. Just click the library button and double click the needed primer for using it for PCR.
For managing the library, open it in the Tools menu. Here you can add a new primer manually or import a set of primers from a file. The added primers are stored between UGENE runs. If you need to share your primers, for example, with your colleagues, you can export them into a file.

