Tutorial: Opening Reading Frame Finder in UGENE
Here we discuss the hottest topics introduced by our users and show the helpful ways of using UGENE, a free cross-platform genome analysis suite.
Today we will continue working with sequence analysis in UGENE and consider searching for open reading frames in a DNA sequence introducing UGENE as an opening reading frame finder. Let's start with opening a nucleotide sequence, for example this file from UGENE package samples. Now we select a sequence region with mouse. To search for open reading frames, we right-click, bringing up the context menu, select „Analyse“ and then „Find open reading frames“ menu items. It will open the UGENE open reading frame finder.
Open Reading Frames Finder Options
The dialog box appears. Let's consider its options. First of all, we can select a strand where to search for open frames, and select the translation code here. The translation code defines, as we see, start codons, alternative start codons, and stop codons. By default UGENE will search for open reading frames that start with one of the „Start codons“, end with one of the „Stop codons“ and their „start codons“ lie within the search range. Also the result open reading frames will be filtered by length, so search result will include only such open reading frames that have length greater than or equal to the value specified in the corresponding text field.
So, we have chosen the translation code. The search range is automatically set to the selected range. It is possible to specify the minimal length the results are filtered by. If the option is unchecked, we will get the results of every length.
Further, checking the „Must terminate within region“ option leads to discarding the results whose stop codons lie beyond the search range. By default the option is unchecked.
Unchecking the next option, „Must start with init codon“, allows such open frames that start with any codon different from a stop codon. The option is checked by default, so we will only get the open reading frames starting one of the „start codons“.
The last option in the parameters panel allows such opening reading frames that start with „Start codons“ or „Alternative start codons“ corresponding to the translation table. By default, only the codons that start with „Start codons“, counts.
Running Search for Open Reading Frames
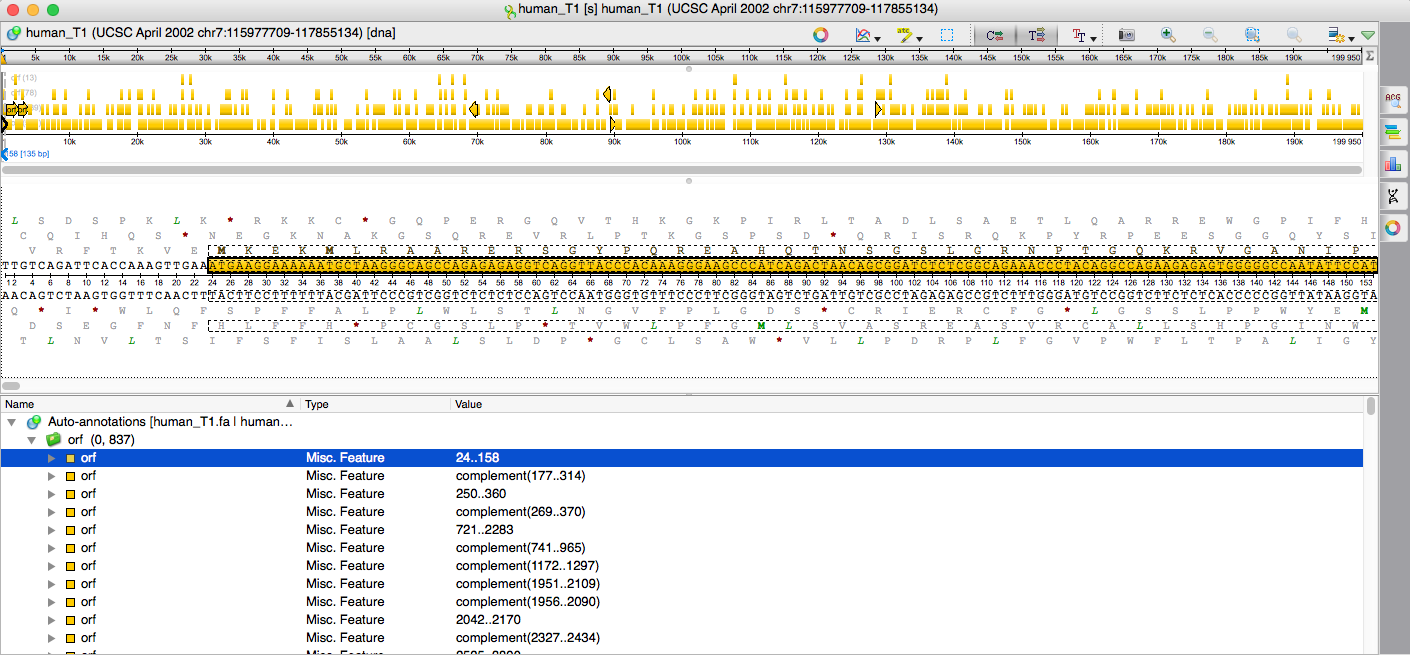
We are ready to perform the search. Click the „Search“ button. The search is done and the resulting opening reading frames are represented as this table rows. It possible to look at the results as the annotations. To do this, we can save the results pressing the „Save as annotations...“ button. In the appeared dialog box we can select the existing or create new annotation table in Genbank format, and specify the results group and annotation names. In our case a new Genbank format file will be created and added to the current project view. Press „Create“.
As we can see, the resulting annotations have appeared at the panoramic and annotations editor views as well.

