UGENE Tips: GC Content, Export Consensus and more
Graphs
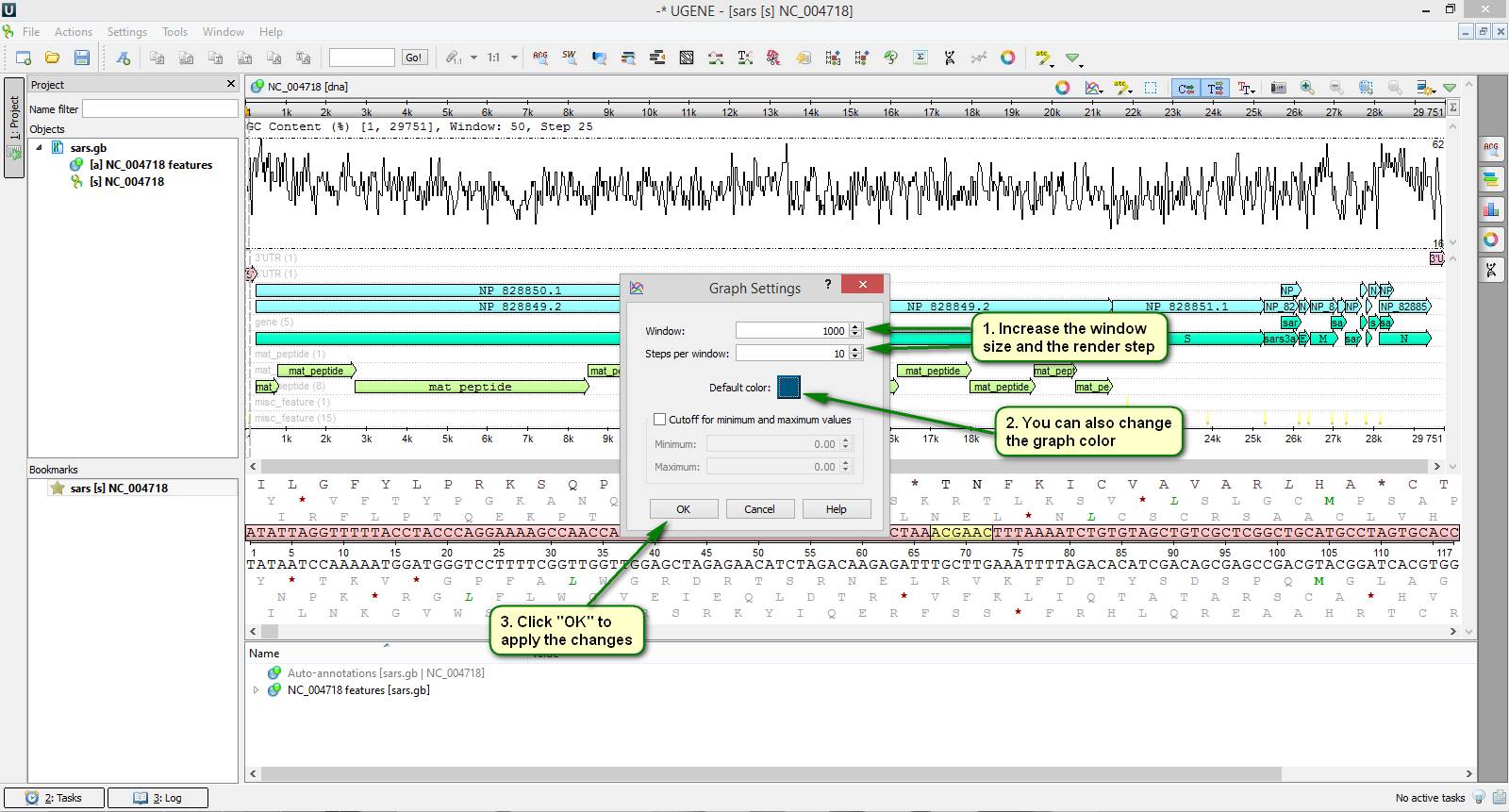
Did you know that you can plot different kinds of statistical graphs based upon your sequences in UGENE?
To view available graph types click the "Graphs" button on the sequence editor toolbar. Then choose the required one from the popup list, for example, "GC Content" or "DNA Flexibility". You can plot several graphs at once for a single sequence. Also graph calculation and rendering settings can be tuned precisely if needed.
You can see an example of how to plot the GC content graph for SARS coronavirus genome on attached screenshots.
See more about working with graphs in the UGENE sequence editor in our documentation.
NCBI Genbank Search
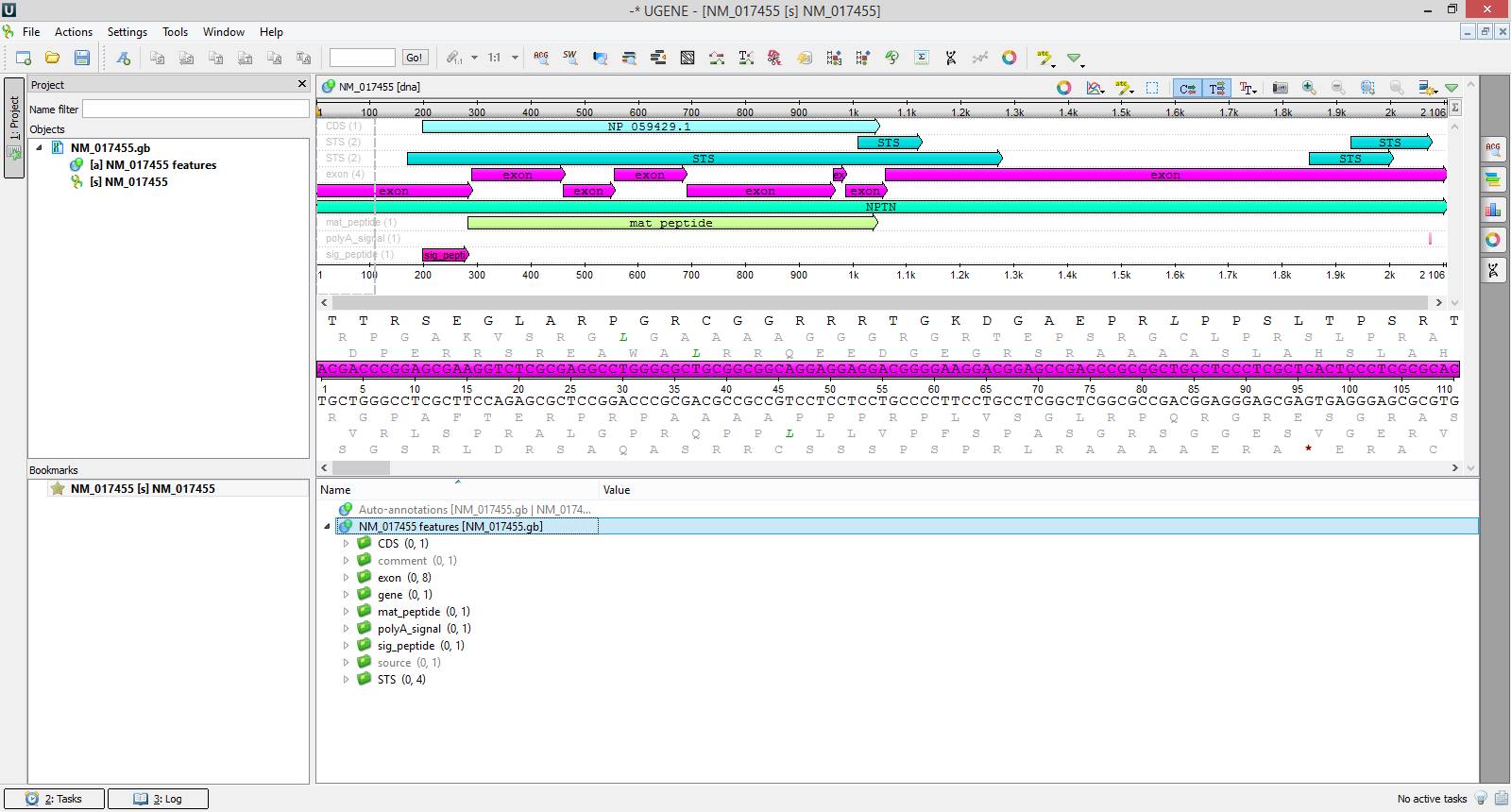
Did you know that you can search sequences in the NCBI Genbank database just from UGENE?
To do this you need to choose { File -> Search NCBI Genbank… } in the main application menu. In the appeared dialog you can make up complex queries including the information about the scientist discovered the sequence, the name of a corresponding gene or organism. After that UGENE provides the list of sequences satisfying specified criteria, and you can choose which of them are to be saved on your computer. By default downloaded sequences are opened immediately in the UGENE sequence editor.
On the attached screenshots you can see an example of how to download the NPTN gene sequence that became known due to the positive influence of its product Neuroplastin on the intelligence.
See more about working with NCBI Genbank in our documentation.
Export Consensus
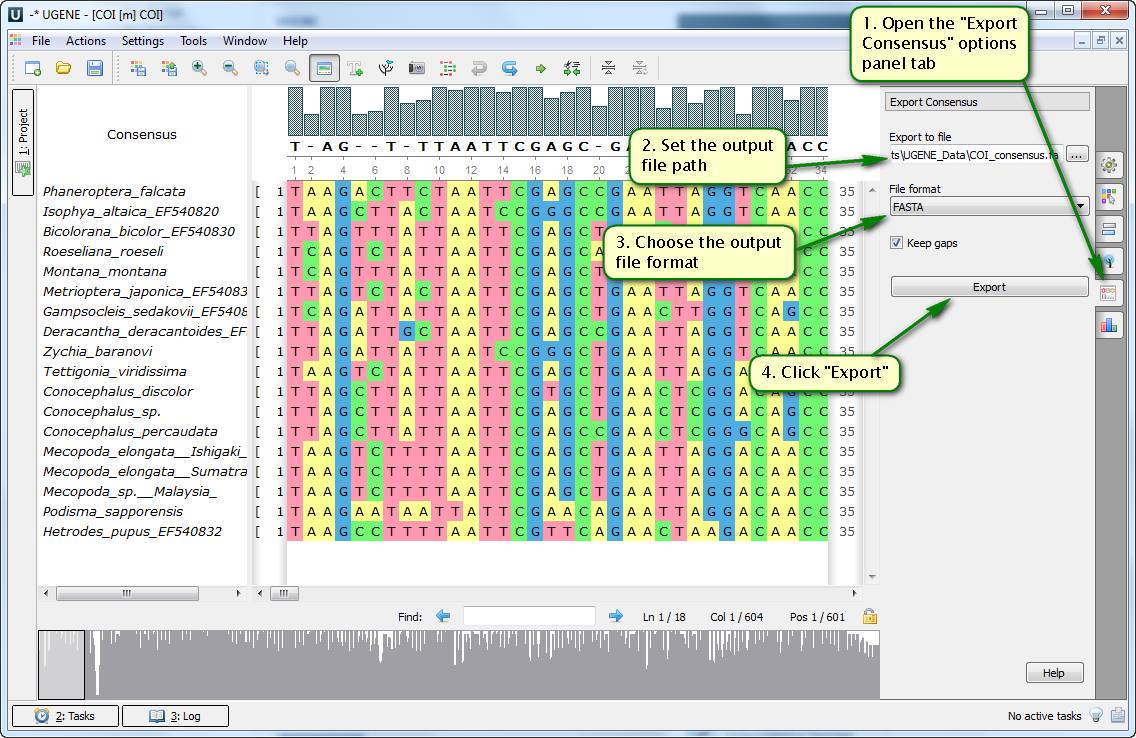
Did you know how to save the consensus sequence of a multiple alignment into a file?
To do this open your file with a multiple alignment in the UGENE Alignment Editor. Then open the "Export Consensus" options panel tab. Here you can specify where the file with the consensus sequence is to be saved to and in which format. Consensus characters calculation algorithm can be set additionally in the "Consensus mode" unit of the "General" options panel tab.
On attached screenshots you can see an example of how to tune and save the consensus sequence of a multiple alignment.
See more about working with the consensus sequence in our documentation.
Find Occurrences

Did you know how to find quickly occurrences of a sequence in a multiple alignment by means of UGENE?
We tried to make it as much easy as it takes from you to find a word on a web-browser page. Therefore, you need just press "Ctrl+F" ("Cmd+F" if you're an OS X user) to activate the input sequence search field, then type your sequence and press "Enter". Successive hits on "Enter" or "Shift+Enter" scroll the multiple alignment to the next or previous found result respectively.
On the attached screenshot you can see an example of how to find the -10 element constituting promoters in prokaryotic cells.
See more about sequence search in the UGENE Multiple Alignment Editor in our documentation.

